Gene
KWMTBOMO00783 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009726
Annotation
translin_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.616
Sequence
CDS
ATGTGCGATAATGAATTGATCAATAAAATATTCAGTGACTTCCAAAAGAATCTCGACCAAGAACAAGAACTACGCGAGACGATTCGGACTATATGTAAAGAGGTTGACCAAATTTCTCGGGAAGCGACCACAGTGCTCCAAGTAATACACTACAATGAAGCCGGCATTGCTCCGGCATGTGGCAAAGCTCGTCTACTTTTCGAGAAGGCCCACGATGGATACGCAAGACTGAAAGATGCTGTACCACCAACCGATTACTTCAAGTATCAGGATCACTGGAGATTCATGACCCAACGCTACTGTTATCTGATAGCGCTCACTATATGGCTAGAGAAGGGAATCCTGGCTTCACACGAGACCATGGCTGAAATATTAGGTGTTAGTCCAGTGGAGTTAAAAGAAGGTTTCCATTTGGACATTGAAGACTATCTGATTGGGCTATTGACGATGTGCTCAGAATTGTCTCGTCTGGCCGTGAACTCGGTGACCCGCGGCGACTACGAGCGCCCCCTGAGGATCTCCAAGTTCGTGATGGAACTGAACGCCGGCTTCAGGCTATTGAACTTGAAGAACGATCATTTGCGCAAACGCTTCGACGCCCTAAAGTACGACGTGAAGAAAATAGAGGAAGTCGTCTACGATCTCAGCATCAGGGGGCTGCTGCCCAAGGGCGATGCGGACCACGGACCAGAACACGAACATAAATAA
Protein
MCDNELINKIFSDFQKNLDQEQELRETIRTICKEVDQISREATTVLQVIHYNEAGIAPACGKARLLFEKAHDGYARLKDAVPPTDYFKYQDHWRFMTQRYCYLIALTIWLEKGILASHETMAEILGVSPVELKEGFHLDIEDYLIGLLTMCSELSRLAVNSVTRGDYERPLRISKFVMELNAGFRLLNLKNDHLRKRFDALKYDVKKIEEVVYDLSIRGLLPKGDADHGPEHEHK
Summary
Uniprot
Q2F5K2
H9JJM5
S4PMD3
A0A194PUP6
A0A194QZB0
A0A2H1WHR0
+ More
A0A1Y1KTW1 A0A182QKR4 A0A182MM13 A0A1W4XBM6 A0A182RN11 A0A067RB54 A0A182WKD1 B0WRW5 Q17D78 A0A1Q3F871 Q16YK8 A0A182HLZ4 A0A182X545 Q7Q729 A0A182VCF5 A0A182NM35 A0A182TR11 A0A182PP51 A0A2M3ZI43 A0A182LJG0 A0A2M4CV01 A0A2M4CV77 A0A182FHX5 A0A182JY27 A0A182J7D9 A0A2J7RQ88 A0A2M3Z8E3 A0A2M3Z8J1 A0A1B0EZ92 A0A182Y107 A0A1B0CQW3 D6WQ79 A0A2M4AKF2 T1E7W8 A0A1L8DDM0 A0A0M4ET37 V5GD89 A0A2M4BYA3 A0A2M4BYJ7 Q28XI6 A0A3B0J098 A0A2P8Z1Y4 U5ESR5 B4GG64 B4P870 A0A0K8TRS2 A0A023EMY0 B3MI45 Q7JVK6 A0A023EKK5 B4KN37 B4MIQ4 B4QAJ9 T1PGB7 B3NN86 A0A1L8DBN3 A0A1I8M9U2 B4J7R1 A0A1W4VPT1 N6TIM3 A0A1L8EBG4 B4LKM0 J3JTG6 A0A0A1XG66 A0A0K8WCU8 A0A034WGZ0 A0A0C9R0J6 A0A1B0AHR0 E2AUJ8 A0A0J7L1R2 A0A1A9WJ07 A0A1I8PZR4 T1JBZ9 A0A0M5J5R7 A0A1A9V5B9 A0A1A9Y2Y9 A0A1B0B080 D3TPR8 A0A0L0BTV3 A0A224YY80 A0A131YF88 A0A336KXM7 A0A154P380 E9G7I9 A0A023FXU5 A0A232EYF1 A0A1E1XCB5 A0A131XPB7 A0A1Z5LEV7 E2C3G6 A0A0P5JV50 A0A0P5NLT6 A0A0N8BSK8 A0A0P5IAX7
A0A1Y1KTW1 A0A182QKR4 A0A182MM13 A0A1W4XBM6 A0A182RN11 A0A067RB54 A0A182WKD1 B0WRW5 Q17D78 A0A1Q3F871 Q16YK8 A0A182HLZ4 A0A182X545 Q7Q729 A0A182VCF5 A0A182NM35 A0A182TR11 A0A182PP51 A0A2M3ZI43 A0A182LJG0 A0A2M4CV01 A0A2M4CV77 A0A182FHX5 A0A182JY27 A0A182J7D9 A0A2J7RQ88 A0A2M3Z8E3 A0A2M3Z8J1 A0A1B0EZ92 A0A182Y107 A0A1B0CQW3 D6WQ79 A0A2M4AKF2 T1E7W8 A0A1L8DDM0 A0A0M4ET37 V5GD89 A0A2M4BYA3 A0A2M4BYJ7 Q28XI6 A0A3B0J098 A0A2P8Z1Y4 U5ESR5 B4GG64 B4P870 A0A0K8TRS2 A0A023EMY0 B3MI45 Q7JVK6 A0A023EKK5 B4KN37 B4MIQ4 B4QAJ9 T1PGB7 B3NN86 A0A1L8DBN3 A0A1I8M9U2 B4J7R1 A0A1W4VPT1 N6TIM3 A0A1L8EBG4 B4LKM0 J3JTG6 A0A0A1XG66 A0A0K8WCU8 A0A034WGZ0 A0A0C9R0J6 A0A1B0AHR0 E2AUJ8 A0A0J7L1R2 A0A1A9WJ07 A0A1I8PZR4 T1JBZ9 A0A0M5J5R7 A0A1A9V5B9 A0A1A9Y2Y9 A0A1B0B080 D3TPR8 A0A0L0BTV3 A0A224YY80 A0A131YF88 A0A336KXM7 A0A154P380 E9G7I9 A0A023FXU5 A0A232EYF1 A0A1E1XCB5 A0A131XPB7 A0A1Z5LEV7 E2C3G6 A0A0P5JV50 A0A0P5NLT6 A0A0N8BSK8 A0A0P5IAX7
Pubmed
19121390
23622113
26354079
28004739
24845553
17510324
+ More
12364791 14747013 17210077 20966253 25244985 18362917 19820115 15632085 17994087 29403074 17550304 26369729 24945155 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18647346 21552261 23650579 26109357 26109356 22936249 25315136 23537049 22516182 25830018 25348373 20798317 20353571 26108605 28797301 26830274 21292972 28648823 28503490 28049606 28528879
12364791 14747013 17210077 20966253 25244985 18362917 19820115 15632085 17994087 29403074 17550304 26369729 24945155 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18647346 21552261 23650579 26109357 26109356 22936249 25315136 23537049 22516182 25830018 25348373 20798317 20353571 26108605 28797301 26830274 21292972 28648823 28503490 28049606 28528879
EMBL
DQ311421
ABD36365.1
BABH01017401
BABH01017402
GAIX01003620
JAA88940.1
+ More
KQ459593 KPI96469.1 KQ460890 KPJ10878.1 ODYU01008722 SOQ52567.1 GEZM01076801 GEZM01076800 GEZM01076799 JAV63640.1 AXCN02002327 AXCM01013109 KK852811 KDR15893.1 DS232061 EDS33530.1 CH477298 EAT44351.1 GFDL01011342 JAV23703.1 CH477515 EAT39699.1 APCN01004280 AAAB01008960 EAA11822.4 GGFM01007430 MBW28181.1 GGFL01004989 MBW69167.1 GGFL01004987 MBW69165.1 NEVH01001345 PNF43004.1 GGFM01004043 MBW24794.1 GGFM01004059 MBW24810.1 AJVK01070341 AJWK01024121 KQ971354 EFA07522.2 GGFK01007891 MBW41212.1 GAMD01002687 JAA98903.1 GFDF01009617 JAV04467.1 CP012524 ALC40612.1 GALX01000331 JAB68135.1 GGFJ01008851 MBW57992.1 GGFJ01008850 MBW57991.1 CM000071 EAL26330.2 OUUW01000001 SPP74115.1 PYGN01000237 PSN50504.1 GANO01002240 JAB57631.1 CH479183 EDW35484.1 CM000158 EDW90117.1 GDAI01001008 JAI16595.1 JXUM01102070 JXUM01111582 GAPW01003889 KQ565518 KQ564697 JAC09709.1 KXJ70906.1 KXJ71879.1 CH902619 EDV38055.1 AE013599 AY118870 AAF58784.1 AAM50730.1 GAPW01003890 JAC09708.1 CH933808 EDW08864.1 CH963719 EDW71993.1 CM000362 CM002911 EDX06504.1 KMY92802.1 KA647852 AFP62481.1 CH954179 EDV56606.1 GFDF01010208 JAV03876.1 CH916367 EDW01117.1 APGK01036455 KB740940 KB632083 ENN77678.1 ERL88621.1 GFDG01002699 JAV16100.1 CH940648 EDW61743.1 BT126522 AEE61486.1 GBXI01004330 JAD09962.1 GDHF01003306 JAI49008.1 GAKP01004121 JAC54831.1 GBYB01006452 JAG76219.1 GL442829 EFN62892.1 LBMM01001376 KMQ96419.1 JH432044 CP012528 ALC48218.1 JXJN01006587 CCAG010001240 EZ423420 ADD19696.1 JRES01001351 KNC23441.1 GFPF01007788 MAA18934.1 GEDV01010524 JAP78033.1 UFQS01001192 UFQT01001192 SSX09620.1 SSX29416.1 KQ434796 KZC05804.1 GL732534 EFX84471.1 GBBL01001805 JAC25515.1 NNAY01001630 OXU23375.1 GFAC01002340 JAT96848.1 GEFH01000593 JAP67988.1 GFJQ02001150 JAW05820.1 GL452320 EFN77504.1 GDIQ01193225 JAK58500.1 GDIQ01150692 JAL01034.1 GDIQ01248163 GDIQ01148995 JAL02731.1 GDIQ01217172 JAK34553.1
KQ459593 KPI96469.1 KQ460890 KPJ10878.1 ODYU01008722 SOQ52567.1 GEZM01076801 GEZM01076800 GEZM01076799 JAV63640.1 AXCN02002327 AXCM01013109 KK852811 KDR15893.1 DS232061 EDS33530.1 CH477298 EAT44351.1 GFDL01011342 JAV23703.1 CH477515 EAT39699.1 APCN01004280 AAAB01008960 EAA11822.4 GGFM01007430 MBW28181.1 GGFL01004989 MBW69167.1 GGFL01004987 MBW69165.1 NEVH01001345 PNF43004.1 GGFM01004043 MBW24794.1 GGFM01004059 MBW24810.1 AJVK01070341 AJWK01024121 KQ971354 EFA07522.2 GGFK01007891 MBW41212.1 GAMD01002687 JAA98903.1 GFDF01009617 JAV04467.1 CP012524 ALC40612.1 GALX01000331 JAB68135.1 GGFJ01008851 MBW57992.1 GGFJ01008850 MBW57991.1 CM000071 EAL26330.2 OUUW01000001 SPP74115.1 PYGN01000237 PSN50504.1 GANO01002240 JAB57631.1 CH479183 EDW35484.1 CM000158 EDW90117.1 GDAI01001008 JAI16595.1 JXUM01102070 JXUM01111582 GAPW01003889 KQ565518 KQ564697 JAC09709.1 KXJ70906.1 KXJ71879.1 CH902619 EDV38055.1 AE013599 AY118870 AAF58784.1 AAM50730.1 GAPW01003890 JAC09708.1 CH933808 EDW08864.1 CH963719 EDW71993.1 CM000362 CM002911 EDX06504.1 KMY92802.1 KA647852 AFP62481.1 CH954179 EDV56606.1 GFDF01010208 JAV03876.1 CH916367 EDW01117.1 APGK01036455 KB740940 KB632083 ENN77678.1 ERL88621.1 GFDG01002699 JAV16100.1 CH940648 EDW61743.1 BT126522 AEE61486.1 GBXI01004330 JAD09962.1 GDHF01003306 JAI49008.1 GAKP01004121 JAC54831.1 GBYB01006452 JAG76219.1 GL442829 EFN62892.1 LBMM01001376 KMQ96419.1 JH432044 CP012528 ALC48218.1 JXJN01006587 CCAG010001240 EZ423420 ADD19696.1 JRES01001351 KNC23441.1 GFPF01007788 MAA18934.1 GEDV01010524 JAP78033.1 UFQS01001192 UFQT01001192 SSX09620.1 SSX29416.1 KQ434796 KZC05804.1 GL732534 EFX84471.1 GBBL01001805 JAC25515.1 NNAY01001630 OXU23375.1 GFAC01002340 JAT96848.1 GEFH01000593 JAP67988.1 GFJQ02001150 JAW05820.1 GL452320 EFN77504.1 GDIQ01193225 JAK58500.1 GDIQ01150692 JAL01034.1 GDIQ01248163 GDIQ01148995 JAL02731.1 GDIQ01217172 JAK34553.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000075886
UP000075883
UP000192223
+ More
UP000075900 UP000027135 UP000075920 UP000002320 UP000008820 UP000075840 UP000076407 UP000007062 UP000075903 UP000075884 UP000075902 UP000075885 UP000075882 UP000069272 UP000075881 UP000075880 UP000235965 UP000092462 UP000076408 UP000092461 UP000007266 UP000092553 UP000001819 UP000268350 UP000245037 UP000008744 UP000002282 UP000069940 UP000249989 UP000007801 UP000000803 UP000009192 UP000007798 UP000000304 UP000008711 UP000095301 UP000001070 UP000192221 UP000019118 UP000030742 UP000008792 UP000092445 UP000000311 UP000036403 UP000091820 UP000095300 UP000078200 UP000092443 UP000092460 UP000092444 UP000037069 UP000076502 UP000000305 UP000215335 UP000008237
UP000075900 UP000027135 UP000075920 UP000002320 UP000008820 UP000075840 UP000076407 UP000007062 UP000075903 UP000075884 UP000075902 UP000075885 UP000075882 UP000069272 UP000075881 UP000075880 UP000235965 UP000092462 UP000076408 UP000092461 UP000007266 UP000092553 UP000001819 UP000268350 UP000245037 UP000008744 UP000002282 UP000069940 UP000249989 UP000007801 UP000000803 UP000009192 UP000007798 UP000000304 UP000008711 UP000095301 UP000001070 UP000192221 UP000019118 UP000030742 UP000008792 UP000092445 UP000000311 UP000036403 UP000091820 UP000095300 UP000078200 UP000092443 UP000092460 UP000092444 UP000037069 UP000076502 UP000000305 UP000215335 UP000008237
Interpro
SUPFAM
SSF74784
SSF74784
Gene 3D
CDD
ProteinModelPortal
Q2F5K2
H9JJM5
S4PMD3
A0A194PUP6
A0A194QZB0
A0A2H1WHR0
+ More
A0A1Y1KTW1 A0A182QKR4 A0A182MM13 A0A1W4XBM6 A0A182RN11 A0A067RB54 A0A182WKD1 B0WRW5 Q17D78 A0A1Q3F871 Q16YK8 A0A182HLZ4 A0A182X545 Q7Q729 A0A182VCF5 A0A182NM35 A0A182TR11 A0A182PP51 A0A2M3ZI43 A0A182LJG0 A0A2M4CV01 A0A2M4CV77 A0A182FHX5 A0A182JY27 A0A182J7D9 A0A2J7RQ88 A0A2M3Z8E3 A0A2M3Z8J1 A0A1B0EZ92 A0A182Y107 A0A1B0CQW3 D6WQ79 A0A2M4AKF2 T1E7W8 A0A1L8DDM0 A0A0M4ET37 V5GD89 A0A2M4BYA3 A0A2M4BYJ7 Q28XI6 A0A3B0J098 A0A2P8Z1Y4 U5ESR5 B4GG64 B4P870 A0A0K8TRS2 A0A023EMY0 B3MI45 Q7JVK6 A0A023EKK5 B4KN37 B4MIQ4 B4QAJ9 T1PGB7 B3NN86 A0A1L8DBN3 A0A1I8M9U2 B4J7R1 A0A1W4VPT1 N6TIM3 A0A1L8EBG4 B4LKM0 J3JTG6 A0A0A1XG66 A0A0K8WCU8 A0A034WGZ0 A0A0C9R0J6 A0A1B0AHR0 E2AUJ8 A0A0J7L1R2 A0A1A9WJ07 A0A1I8PZR4 T1JBZ9 A0A0M5J5R7 A0A1A9V5B9 A0A1A9Y2Y9 A0A1B0B080 D3TPR8 A0A0L0BTV3 A0A224YY80 A0A131YF88 A0A336KXM7 A0A154P380 E9G7I9 A0A023FXU5 A0A232EYF1 A0A1E1XCB5 A0A131XPB7 A0A1Z5LEV7 E2C3G6 A0A0P5JV50 A0A0P5NLT6 A0A0N8BSK8 A0A0P5IAX7
A0A1Y1KTW1 A0A182QKR4 A0A182MM13 A0A1W4XBM6 A0A182RN11 A0A067RB54 A0A182WKD1 B0WRW5 Q17D78 A0A1Q3F871 Q16YK8 A0A182HLZ4 A0A182X545 Q7Q729 A0A182VCF5 A0A182NM35 A0A182TR11 A0A182PP51 A0A2M3ZI43 A0A182LJG0 A0A2M4CV01 A0A2M4CV77 A0A182FHX5 A0A182JY27 A0A182J7D9 A0A2J7RQ88 A0A2M3Z8E3 A0A2M3Z8J1 A0A1B0EZ92 A0A182Y107 A0A1B0CQW3 D6WQ79 A0A2M4AKF2 T1E7W8 A0A1L8DDM0 A0A0M4ET37 V5GD89 A0A2M4BYA3 A0A2M4BYJ7 Q28XI6 A0A3B0J098 A0A2P8Z1Y4 U5ESR5 B4GG64 B4P870 A0A0K8TRS2 A0A023EMY0 B3MI45 Q7JVK6 A0A023EKK5 B4KN37 B4MIQ4 B4QAJ9 T1PGB7 B3NN86 A0A1L8DBN3 A0A1I8M9U2 B4J7R1 A0A1W4VPT1 N6TIM3 A0A1L8EBG4 B4LKM0 J3JTG6 A0A0A1XG66 A0A0K8WCU8 A0A034WGZ0 A0A0C9R0J6 A0A1B0AHR0 E2AUJ8 A0A0J7L1R2 A0A1A9WJ07 A0A1I8PZR4 T1JBZ9 A0A0M5J5R7 A0A1A9V5B9 A0A1A9Y2Y9 A0A1B0B080 D3TPR8 A0A0L0BTV3 A0A224YY80 A0A131YF88 A0A336KXM7 A0A154P380 E9G7I9 A0A023FXU5 A0A232EYF1 A0A1E1XCB5 A0A131XPB7 A0A1Z5LEV7 E2C3G6 A0A0P5JV50 A0A0P5NLT6 A0A0N8BSK8 A0A0P5IAX7
PDB
4DG7
E-value=1.34974e-68,
Score=657
Ontologies
GO
PANTHER
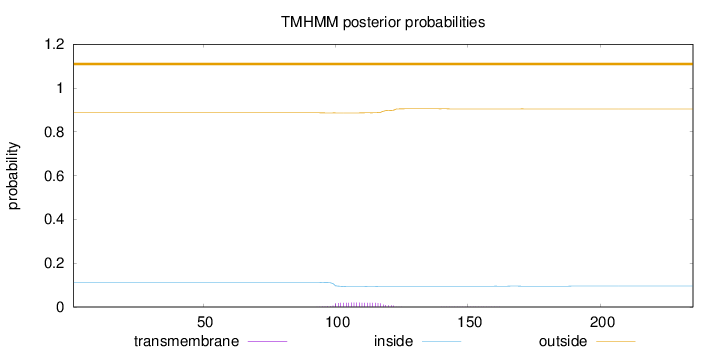
Topology
Length:
235
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.41176
Exp number, first 60 AAs:
0.00061
Total prob of N-in:
0.11305
outside
1 - 235
Population Genetic Test Statistics
Pi
250.432925
Theta
184.797214
Tajima's D
0.981214
CLR
0.087392
CSRT
0.656867156642168
Interpretation
Uncertain
