Gene
KWMTBOMO00776
Pre Gene Modal
BGIBMGA009708
Annotation
origin_recognition_complex_subunit_4_[Bombyx_mori]
Full name
Origin recognition complex subunit 4
Location in the cell
Nuclear Reliability : 3.23
Sequence
CDS
ATGGATACAGACCCGTTGGCTATTCAAATAAAACTTGTAAGGAAGTACATAAAATTGAAATTAATAGAAGACAATGTATTGTTTCGGGGGCACGACCAGGAAAGAAATCATGTGTACGAATTATTAAGAAGAACTGTCTTACAAGGAGAATCGCATTCGGCTTTACTTATAGGGCCGAGAAGTAGTGGAAAAACCACTCTACTAAACTCGGTACTTCACCAGCTCTCTAGAGAGACAGATTTGGAGAATGATGCTATTATAATTCAACTGAATGGCCTTGTACATAGTGATGATAAACTGGCCCTTAAAGCTATCACAGCGCAGATGCAATTAGAAAATACTGTGGGTGAACGAGTTTTCGGTACCTTTGCCGAGAACCTCTCGTTTTTGTTAAGCTGCATCCAGACTGGTGTTGACAGACGATGTCGAAGCATGGTATTCATTCTAGACCAGTGTGACATGTTTTGCCACAGCGGTATCTCCCAGACATTACTGTATAACTTATTTGACGTAACTCATTCACAGACGGCTCCTATGTGTGTTATCGGTGTAACAAATCGCTTAGACATTATGGAGTTGTTTGAAAAAAGGGTCAAATCCCGTTTTTCCCATCGTCACATATTCATATTTCCTAATGAAGTCAATGATGATGACCGGTATGATATTATCTCGCCTCTGGAAGGGAGAAAGAGATTGTTTGTGGAATTGCTGAGTCTGCCCCTGGAGGGTTCTGTGTCGCCGAGGAAGAAAAGAAAATCAAGGAAGTCCATCGCCAAAACTGATGAAACGGCCTGCACATCCACAGTCTTTGCAATACCAGAACATATATTAAAAGGGAAACTCGACGAGCAATGTTTGTCTACGATGGACCGAGCCTTTGTGAGCGACTGGAATGCCCATATCAGAAGTTTGATTGAAAACGAAAAAGTCATTGACGCTTTAGAGAAGCTATGTTACTATACCGTTGATGAGCAAACTTTTAGGAATGTTTTGTTTGAAACAATCAGTAAACTGTCTCCGACGAAACCGAATATAAACGTCATAGATCTAACAACGGCCATAGACAACGCAGTGGCTCCGGAGCACAGAGTCAAAGTGTTGCAGTCGCTATCTATACTAGAGTTGTCTCTGGTGATAGCCATGAAGCACGGGATGGAGATATTCGACGGGCAACCAATGAATTTTGAAATGATCCTACACAGGTACACGAAGTTCGCAAACGAGCACTCCTCGTCGCAGAGCGTGCCTCGCCCCGTCATCCTCAAGGCGTTCGAGCACCTGCAGGCGCTGGAGATCGTGTTGCCGGCCAGAACGTCGGATGTCGGGGGCGCGGCGGGCGACGCCTCCTCCAGCCGCGTGCAGAAGGAATACAAGCTGTACACGCTGGCCGTGCCCGCCGAGGACCTGGGACGAGCCGTCGACACCTTCAAGAGTCTACCCACCGAGATCTGCCATTGGTACAGCAATTCCGTCGTGTGA
Protein
MDTDPLAIQIKLVRKYIKLKLIEDNVLFRGHDQERNHVYELLRRTVLQGESHSALLIGPRSSGKTTLLNSVLHQLSRETDLENDAIIIQLNGLVHSDDKLALKAITAQMQLENTVGERVFGTFAENLSFLLSCIQTGVDRRCRSMVFILDQCDMFCHSGISQTLLYNLFDVTHSQTAPMCVIGVTNRLDIMELFEKRVKSRFSHRHIFIFPNEVNDDDRYDIISPLEGRKRLFVELLSLPLEGSVSPRKKRKSRKSIAKTDETACTSTVFAIPEHILKGKLDEQCLSTMDRAFVSDWNAHIRSLIENEKVIDALEKLCYYTVDEQTFRNVLFETISKLSPTKPNINVIDLTTAIDNAVAPEHRVKVLQSLSILELSLVIAMKHGMEIFDGQPMNFEMILHRYTKFANEHSSSQSVPRPVILKAFEHLQALEIVLPARTSDVGGAAGDASSSRVQKEYKLYTLAVPAEDLGRAVDTFKSLPTEICHWYSNSVV
Summary
Description
Component of the origin recognition complex (ORC) that binds origins of replication.
Similarity
Belongs to the ORC4 family.
Uniprot
D3VW59
H9JJK7
A0A194PZ94
A0A212EK47
A0A2H1VMX4
A0A194R024
+ More
A0A2A4IX77 A0A310S9U0 A0A0M9A8V5 A0A151IIS9 A0A151WZ50 A0A0C9R9M2 A0A158P2Y5 E2AUK1 E9ICQ5 A0A2A3EIX8 A0A0L7RCY8 A0A088AM82 A0A195DND8 F4X5H2 A0A182SQ06 A0A182M6A9 A0A182WL02 B0WCN1 A0A232FBN1 A0A1Q3F134 K7J3C9 A0A182GME7 A0A195BPW2 Q17K74 A0A1S4EZY3 A0A182NJQ2 A0A067QT57 A0A182RMZ4 A0A182Y259 A0A3L8DRX1 A0A084WJQ9 A0A1I8NVM2 A0A182FEQ6 E2C3G2 W5JUX6 A0A182JRQ1 A0A182WWT4 A0A0L0BPH8 A0A182L370 A0A182TFQ6 Q7QIT4 A0A1A9WBB4 A0A1I8NG94 A0A182PHZ6 A0A182VNT2 A0A1A9UY68 Q28Z26 B4GID3 A0A1B0A7I8 A0A195EZA0 A0A182Q574 A0A1B0BB17 B4J4C9 A0A0M4EF56 A0A3B0J4P4 A0A0K8U8C7 A0A1A9XN59 B4KTJ5 A0A034WT52 B4LN60 B4MPA3 A0A1W4V061 A0A0A1WH20 W8C2W6 B3MC30 A0A1B0GG68 A0A0J7KLR2 B3NQZ3 A0A0J9RKS5 B4PBV5 B4IH88 D6WQ61 A0A0K8TT39 A0A026WX24 Q9W102 Q9Y1B3 N6TUL5 U4UPU4 A0A182HWG3 A0A1J1ISJ5 A0A023F1E5 A0A182J7M8 A0A210QBF1 A0A336MTH8 A0A1Y1LQ05 A0A1E1XSK2 A0A293LKI2 R7VC25 A0A1S3GZ31 A0A1S3GYH9 M4A861 A0A131YN83 A0A1E1XBB2 W5LZL4
A0A2A4IX77 A0A310S9U0 A0A0M9A8V5 A0A151IIS9 A0A151WZ50 A0A0C9R9M2 A0A158P2Y5 E2AUK1 E9ICQ5 A0A2A3EIX8 A0A0L7RCY8 A0A088AM82 A0A195DND8 F4X5H2 A0A182SQ06 A0A182M6A9 A0A182WL02 B0WCN1 A0A232FBN1 A0A1Q3F134 K7J3C9 A0A182GME7 A0A195BPW2 Q17K74 A0A1S4EZY3 A0A182NJQ2 A0A067QT57 A0A182RMZ4 A0A182Y259 A0A3L8DRX1 A0A084WJQ9 A0A1I8NVM2 A0A182FEQ6 E2C3G2 W5JUX6 A0A182JRQ1 A0A182WWT4 A0A0L0BPH8 A0A182L370 A0A182TFQ6 Q7QIT4 A0A1A9WBB4 A0A1I8NG94 A0A182PHZ6 A0A182VNT2 A0A1A9UY68 Q28Z26 B4GID3 A0A1B0A7I8 A0A195EZA0 A0A182Q574 A0A1B0BB17 B4J4C9 A0A0M4EF56 A0A3B0J4P4 A0A0K8U8C7 A0A1A9XN59 B4KTJ5 A0A034WT52 B4LN60 B4MPA3 A0A1W4V061 A0A0A1WH20 W8C2W6 B3MC30 A0A1B0GG68 A0A0J7KLR2 B3NQZ3 A0A0J9RKS5 B4PBV5 B4IH88 D6WQ61 A0A0K8TT39 A0A026WX24 Q9W102 Q9Y1B3 N6TUL5 U4UPU4 A0A182HWG3 A0A1J1ISJ5 A0A023F1E5 A0A182J7M8 A0A210QBF1 A0A336MTH8 A0A1Y1LQ05 A0A1E1XSK2 A0A293LKI2 R7VC25 A0A1S3GZ31 A0A1S3GYH9 M4A861 A0A131YN83 A0A1E1XBB2 W5LZL4
Pubmed
21162711
19121390
26354079
22118469
21347285
20798317
+ More
21282665 21719571 28648823 20075255 26483478 17510324 24845553 25244985 30249741 24438588 20920257 23761445 26108605 20966253 12364791 14747013 17210077 25315136 15632085 17994087 25348373 25830018 24495485 22936249 17550304 18362917 19820115 26369729 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25762138 10346817 23537049 25474469 28812685 28004739 29209593 23254933 23542700 26830274 28503490
21282665 21719571 28648823 20075255 26483478 17510324 24845553 25244985 30249741 24438588 20920257 23761445 26108605 20966253 12364791 14747013 17210077 25315136 15632085 17994087 25348373 25830018 24495485 22936249 17550304 18362917 19820115 26369729 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25762138 10346817 23537049 25474469 28812685 28004739 29209593 23254933 23542700 26830274 28503490
EMBL
GQ214393
ADD10140.1
BABH01017393
BABH01017394
KQ459593
KPI96475.1
+ More
AGBW02014329 OWR41856.1 ODYU01003411 SOQ42146.1 KQ460890 KPJ10884.1 NWSH01005380 PCG64259.1 KQ762882 OAD55373.1 KQ435711 KOX79615.1 KQ977459 KYN02567.1 KQ982649 KYQ52981.1 GBYB01003362 GBYB01003363 GBYB01004880 JAG73129.1 JAG73130.1 JAG74647.1 ADTU01007645 GL442829 EFN62895.1 GL762354 EFZ21642.1 KZ288248 PBC31139.1 KQ414615 KOC68679.1 KQ980713 KYN14383.1 GL888716 EGI58328.1 AXCM01004892 DS231889 EDS43682.1 NNAY01000520 OXU27880.1 GFDL01013806 JAV21239.1 JXUM01014086 KQ560393 KXJ82659.1 KQ976432 KYM87532.1 CH477227 EAT47112.1 KK853050 KDR12067.1 QOIP01000005 RLU23154.1 ATLV01024046 KE525348 KFB50453.1 GL452320 EFN77500.1 ADMH02000419 ETN66574.1 JRES01001567 KNC21906.1 AAAB01008807 EAA03944.2 CM000071 EAL25789.1 CH479183 EDW36253.1 KQ981905 KYN33456.1 AXCN02000514 JXJN01011276 CH916367 EDW01611.1 CP012524 ALC41914.1 OUUW01000001 SPP74513.1 GDHF01029481 JAI22833.1 CH933808 EDW08556.1 GAKP01001128 JAC57824.1 CH940648 EDW60064.1 CH963849 EDW73942.1 GBXI01016003 JAC98288.1 GAMC01002987 JAC03569.1 CH902619 EDV37217.1 CCAG010022550 LBMM01005645 KMQ91338.1 CH954179 EDV57076.1 CM002911 KMY96441.1 CM000158 EDW92609.1 CH480838 EDW49264.1 KQ971354 EFA06929.1 GDAI01000290 JAI17313.1 KK107099 EZA59679.1 AE013599 BT004868 AAF47276.1 AAO45224.1 AF139063 AAD39473.1 APGK01051641 KB741198 ENN72970.1 KB632313 ERL92166.1 APCN01001490 CVRI01000059 CRL03104.1 GBBI01003898 JAC14814.1 NEDP02004288 OWF46063.1 UFQT01002418 SSX33410.1 GEZM01049963 JAV75729.1 GFAA01001164 JAU02271.1 GFWV01002590 MAA27320.1 AMQN01000610 KB293180 ELU16403.1 GEDV01008667 JAP79890.1 GFAC01002653 JAT96535.1 AHAT01011655
AGBW02014329 OWR41856.1 ODYU01003411 SOQ42146.1 KQ460890 KPJ10884.1 NWSH01005380 PCG64259.1 KQ762882 OAD55373.1 KQ435711 KOX79615.1 KQ977459 KYN02567.1 KQ982649 KYQ52981.1 GBYB01003362 GBYB01003363 GBYB01004880 JAG73129.1 JAG73130.1 JAG74647.1 ADTU01007645 GL442829 EFN62895.1 GL762354 EFZ21642.1 KZ288248 PBC31139.1 KQ414615 KOC68679.1 KQ980713 KYN14383.1 GL888716 EGI58328.1 AXCM01004892 DS231889 EDS43682.1 NNAY01000520 OXU27880.1 GFDL01013806 JAV21239.1 JXUM01014086 KQ560393 KXJ82659.1 KQ976432 KYM87532.1 CH477227 EAT47112.1 KK853050 KDR12067.1 QOIP01000005 RLU23154.1 ATLV01024046 KE525348 KFB50453.1 GL452320 EFN77500.1 ADMH02000419 ETN66574.1 JRES01001567 KNC21906.1 AAAB01008807 EAA03944.2 CM000071 EAL25789.1 CH479183 EDW36253.1 KQ981905 KYN33456.1 AXCN02000514 JXJN01011276 CH916367 EDW01611.1 CP012524 ALC41914.1 OUUW01000001 SPP74513.1 GDHF01029481 JAI22833.1 CH933808 EDW08556.1 GAKP01001128 JAC57824.1 CH940648 EDW60064.1 CH963849 EDW73942.1 GBXI01016003 JAC98288.1 GAMC01002987 JAC03569.1 CH902619 EDV37217.1 CCAG010022550 LBMM01005645 KMQ91338.1 CH954179 EDV57076.1 CM002911 KMY96441.1 CM000158 EDW92609.1 CH480838 EDW49264.1 KQ971354 EFA06929.1 GDAI01000290 JAI17313.1 KK107099 EZA59679.1 AE013599 BT004868 AAF47276.1 AAO45224.1 AF139063 AAD39473.1 APGK01051641 KB741198 ENN72970.1 KB632313 ERL92166.1 APCN01001490 CVRI01000059 CRL03104.1 GBBI01003898 JAC14814.1 NEDP02004288 OWF46063.1 UFQT01002418 SSX33410.1 GEZM01049963 JAV75729.1 GFAA01001164 JAU02271.1 GFWV01002590 MAA27320.1 AMQN01000610 KB293180 ELU16403.1 GEDV01008667 JAP79890.1 GFAC01002653 JAT96535.1 AHAT01011655
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000218220
UP000053105
+ More
UP000078542 UP000075809 UP000005205 UP000000311 UP000242457 UP000053825 UP000005203 UP000078492 UP000007755 UP000075901 UP000075883 UP000075920 UP000002320 UP000215335 UP000002358 UP000069940 UP000249989 UP000078540 UP000008820 UP000075884 UP000027135 UP000075900 UP000076408 UP000279307 UP000030765 UP000095300 UP000069272 UP000008237 UP000000673 UP000075881 UP000076407 UP000037069 UP000075882 UP000075902 UP000007062 UP000091820 UP000095301 UP000075885 UP000075903 UP000078200 UP000001819 UP000008744 UP000092445 UP000078541 UP000075886 UP000092460 UP000001070 UP000092553 UP000268350 UP000092443 UP000009192 UP000008792 UP000007798 UP000192221 UP000007801 UP000092444 UP000036403 UP000008711 UP000002282 UP000001292 UP000007266 UP000053097 UP000000803 UP000019118 UP000030742 UP000075840 UP000183832 UP000075880 UP000242188 UP000014760 UP000085678 UP000002852 UP000018468
UP000078542 UP000075809 UP000005205 UP000000311 UP000242457 UP000053825 UP000005203 UP000078492 UP000007755 UP000075901 UP000075883 UP000075920 UP000002320 UP000215335 UP000002358 UP000069940 UP000249989 UP000078540 UP000008820 UP000075884 UP000027135 UP000075900 UP000076408 UP000279307 UP000030765 UP000095300 UP000069272 UP000008237 UP000000673 UP000075881 UP000076407 UP000037069 UP000075882 UP000075902 UP000007062 UP000091820 UP000095301 UP000075885 UP000075903 UP000078200 UP000001819 UP000008744 UP000092445 UP000078541 UP000075886 UP000092460 UP000001070 UP000092553 UP000268350 UP000092443 UP000009192 UP000008792 UP000007798 UP000192221 UP000007801 UP000092444 UP000036403 UP000008711 UP000002282 UP000001292 UP000007266 UP000053097 UP000000803 UP000019118 UP000030742 UP000075840 UP000183832 UP000075880 UP000242188 UP000014760 UP000085678 UP000002852 UP000018468
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
D3VW59
H9JJK7
A0A194PZ94
A0A212EK47
A0A2H1VMX4
A0A194R024
+ More
A0A2A4IX77 A0A310S9U0 A0A0M9A8V5 A0A151IIS9 A0A151WZ50 A0A0C9R9M2 A0A158P2Y5 E2AUK1 E9ICQ5 A0A2A3EIX8 A0A0L7RCY8 A0A088AM82 A0A195DND8 F4X5H2 A0A182SQ06 A0A182M6A9 A0A182WL02 B0WCN1 A0A232FBN1 A0A1Q3F134 K7J3C9 A0A182GME7 A0A195BPW2 Q17K74 A0A1S4EZY3 A0A182NJQ2 A0A067QT57 A0A182RMZ4 A0A182Y259 A0A3L8DRX1 A0A084WJQ9 A0A1I8NVM2 A0A182FEQ6 E2C3G2 W5JUX6 A0A182JRQ1 A0A182WWT4 A0A0L0BPH8 A0A182L370 A0A182TFQ6 Q7QIT4 A0A1A9WBB4 A0A1I8NG94 A0A182PHZ6 A0A182VNT2 A0A1A9UY68 Q28Z26 B4GID3 A0A1B0A7I8 A0A195EZA0 A0A182Q574 A0A1B0BB17 B4J4C9 A0A0M4EF56 A0A3B0J4P4 A0A0K8U8C7 A0A1A9XN59 B4KTJ5 A0A034WT52 B4LN60 B4MPA3 A0A1W4V061 A0A0A1WH20 W8C2W6 B3MC30 A0A1B0GG68 A0A0J7KLR2 B3NQZ3 A0A0J9RKS5 B4PBV5 B4IH88 D6WQ61 A0A0K8TT39 A0A026WX24 Q9W102 Q9Y1B3 N6TUL5 U4UPU4 A0A182HWG3 A0A1J1ISJ5 A0A023F1E5 A0A182J7M8 A0A210QBF1 A0A336MTH8 A0A1Y1LQ05 A0A1E1XSK2 A0A293LKI2 R7VC25 A0A1S3GZ31 A0A1S3GYH9 M4A861 A0A131YN83 A0A1E1XBB2 W5LZL4
A0A2A4IX77 A0A310S9U0 A0A0M9A8V5 A0A151IIS9 A0A151WZ50 A0A0C9R9M2 A0A158P2Y5 E2AUK1 E9ICQ5 A0A2A3EIX8 A0A0L7RCY8 A0A088AM82 A0A195DND8 F4X5H2 A0A182SQ06 A0A182M6A9 A0A182WL02 B0WCN1 A0A232FBN1 A0A1Q3F134 K7J3C9 A0A182GME7 A0A195BPW2 Q17K74 A0A1S4EZY3 A0A182NJQ2 A0A067QT57 A0A182RMZ4 A0A182Y259 A0A3L8DRX1 A0A084WJQ9 A0A1I8NVM2 A0A182FEQ6 E2C3G2 W5JUX6 A0A182JRQ1 A0A182WWT4 A0A0L0BPH8 A0A182L370 A0A182TFQ6 Q7QIT4 A0A1A9WBB4 A0A1I8NG94 A0A182PHZ6 A0A182VNT2 A0A1A9UY68 Q28Z26 B4GID3 A0A1B0A7I8 A0A195EZA0 A0A182Q574 A0A1B0BB17 B4J4C9 A0A0M4EF56 A0A3B0J4P4 A0A0K8U8C7 A0A1A9XN59 B4KTJ5 A0A034WT52 B4LN60 B4MPA3 A0A1W4V061 A0A0A1WH20 W8C2W6 B3MC30 A0A1B0GG68 A0A0J7KLR2 B3NQZ3 A0A0J9RKS5 B4PBV5 B4IH88 D6WQ61 A0A0K8TT39 A0A026WX24 Q9W102 Q9Y1B3 N6TUL5 U4UPU4 A0A182HWG3 A0A1J1ISJ5 A0A023F1E5 A0A182J7M8 A0A210QBF1 A0A336MTH8 A0A1Y1LQ05 A0A1E1XSK2 A0A293LKI2 R7VC25 A0A1S3GZ31 A0A1S3GYH9 M4A861 A0A131YN83 A0A1E1XBB2 W5LZL4
PDB
4XGC
E-value=6.26877e-86,
Score=810
Ontologies
GO
PANTHER
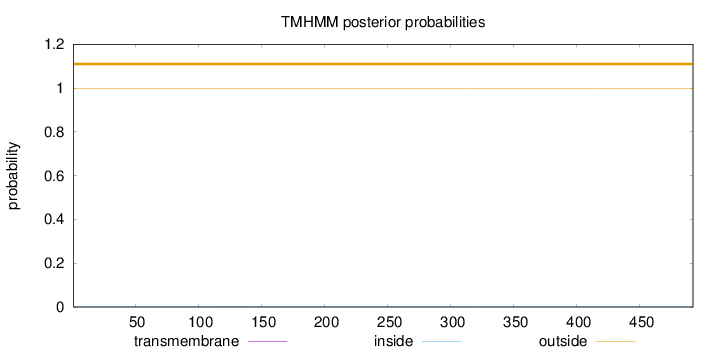
Topology
Subcellular location
Nucleus
Length:
492
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01682
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00227
outside
1 - 492
Population Genetic Test Statistics
Pi
277.931103
Theta
163.897132
Tajima's D
1.301586
CLR
0.014322
CSRT
0.734113294335283
Interpretation
Uncertain
