Gene
KWMTBOMO00775
Pre Gene Modal
BGIBMGA009724
Annotation
PREDICTED:_putative_ammonium_transporter_2_[Papilio_machaon]
Full name
Ammonium transporter
Location in the cell
PlasmaMembrane Reliability : 3.597
Sequence
CDS
ATGGATCCGAACTGGACCCACGTTGGACTTCGGTCACCTAACCTCACGAACACTACAGTGAACCTTCAGGAGAACTTCGAGATCGACTTAGAAGATACAAACTGGATTTTGACTTCGTCTTTCATCATCTTCACAATGCAGACTGGTTTCGGCATGTTGGAATCCGGTTGCGTTTCCATTAAGAACGAGGCCAACATTATGATGAAGAATATGGCTGATATATCGCTCGGGGGTCTCACCTACTGGATATTCGGTTACGGGCTCTCATTCGGCGAGAGCTCCTACTCCAATCCGTTCGTTGGTACTGGAGACTTCCTACTTGATCCTCCAGTGGGAGACGCGCTGATGGGTCCAGTGTTCGCTTCGTTTCTCTTCCAGCTGTCCTTCGCCACCACTGCGACTACTATTGTCAGTGGCGCGATGGCTGAACGGTGTAACTTCAAAGCGTACTGCGTTTTCTCGTTTCTAAACACGATCGTGTACTGCGTCCCAGCCGGCTGGGTGTGGGGCTCGCATGGCTTCCTCAACCAGCTCGGTGCTGTGGACATCGCCGGCTCCGGCCCTGTGCACCTTGTGGGTGGATCGTCAGCTTTTGCGTCAGCTTTGATGCTGGGACCGAGACTGGGCAGGTATGCCCGGGGCACGGACCCTTTACCCCTCGGCAACCCGGTCAACGCCGCCATGGGAACGTTCGTGCTTTGGTGGGGATGGTTGGCTTTCAACTCCGGCAGCACATATGGGGTGAGCGGTGCCAAGTGGCAGTACGCAGCGAGAGCGGCCGTCATGACCATGATGGGTTCTTTCGGTGGTGGATGCTTCGGTCTACTGTTCACGCTTATCAAGAACAAAGGAAAAGCTGAGGTTATGGAGCTGATTAACAGCATCCTCGGTTCTCTGGTGTCTATAACAGCTGGCTGCTTCCTGTACCGCGCTTGGGAGTCTCTGCTGATAGGATTCATTGGTGCCGGCGTGGCTTCGGCGAGCTCGATGCTATTCGACAGACTCCGGGTCGACGACCCCGTGGGAGCTTCGGCTGTCCACGGCGCTTGCGGAATCTGGGGAGTCATAGCAGTTGGTCTATTTGCCGACAACCCTATACCGATGGAGACGACAAACGGGAGATCCGGCTTGTTCAAAGGTGGCGGCTGGTACCTCCTGGGAGTGCAGAGCCTCACCGCCGTGTGCCTGGCCGCGTGGGGCGTGCTGGTCACCATGCTGTTACTGTGGCTCATAGACAGGGTCATGCCTATACGAATGGACCCTTACAATGAGTTGCTTGGGGCCGACCTTACTGAGCACAGGATCAGGCATGGACAGGTGGGAGTGTCTCGCGCTGTGTCAGCCCTGAGGCCGTTCCGCCGCTCAAGCAGCATGGAGGAAGTCGGGACTATCGGCATCAACACCGGACACGGTCTGGTCGTCGAAAAAATCCTCAAGGCGAACAAACGTAACGAGCTCGATGGGAAGCCGCAAACCTACACGAACGAAGCGTTCGTCGCCGCAGACAGCGCCTCAACGTCGTACAAAAACAACGGACTCCTGAACAAGCGTCCCAATGGCAACGGAGACCAGCTCCACCGCACTCTAGACTCTATGATCGATGAGATTAATGGCGGGAAACGCGAAACGTCAAAAGGCGCGCGAAAAAACAATGGCGTCCCGGTGATACCAGACGAAATAAAAGGAAAAAAATTTAGGGAGTTAACGGTGACCGAACTGGATGAGTTGGGCGAAATTAATGCCGCCCAGGATAGGTTCCGGCGGAGATTCGGCGAGGAGGATTACAAGAGTGACGTTATGAGGAGTGCAACGATAACTCCTAATGTGCGGTGGATTGATTGA
Protein
MDPNWTHVGLRSPNLTNTTVNLQENFEIDLEDTNWILTSSFIIFTMQTGFGMLESGCVSIKNEANIMMKNMADISLGGLTYWIFGYGLSFGESSYSNPFVGTGDFLLDPPVGDALMGPVFASFLFQLSFATTATTIVSGAMAERCNFKAYCVFSFLNTIVYCVPAGWVWGSHGFLNQLGAVDIAGSGPVHLVGGSSAFASALMLGPRLGRYARGTDPLPLGNPVNAAMGTFVLWWGWLAFNSGSTYGVSGAKWQYAARAAVMTMMGSFGGGCFGLLFTLIKNKGKAEVMELINSILGSLVSITAGCFLYRAWESLLIGFIGAGVASASSMLFDRLRVDDPVGASAVHGACGIWGVIAVGLFADNPIPMETTNGRSGLFKGGGWYLLGVQSLTAVCLAAWGVLVTMLLLWLIDRVMPIRMDPYNELLGADLTEHRIRHGQVGVSRAVSALRPFRRSSSMEEVGTIGINTGHGLVVEKILKANKRNELDGKPQTYTNEAFVAADSASTSYKNNGLLNKRPNGNGDQLHRTLDSMIDEINGGKRETSKGARKNNGVPVIPDEIKGKKFRELTVTELDELGEINAAQDRFRRRFGEEDYKSDVMRSATITPNVRWID
Summary
Similarity
Belongs to the ammonia transporter channel (TC 1.A.11.2) family.
Uniprot
H9JJM3
A0A194R0V9
A0A194PSR8
A0A2A4IXX3
A0A2H1VMS8
B4K5V9
+ More
B4N8M1 A0A0M3QYI4 A0A1S4FGF9 A0A1I8P675 Q9VFA9 B4LZZ8 B4HDZ9 B4JGC0 Q294C0 A0A1W4VMK1 A0A1A9ZY94 A0A1A9XWD0 A0A3B0KCL2 A0A034WGQ2 W8BZ14 A0A1B0BL43 A0A3B0KJK5 A0A1A9VAX2 B3MX93 B4PS70 A0A0R1E419 B3P0S7 A0A067QRF1 A0A0L0BPN5 A0A2J7QL05 A0A084W5B9 A0A182ITC2 A0A139WEF3 A0A182PF20 A0A182F835 A0A139WE35 A0A1Y1KLR8 A0A336MI61 A0A336MBT3 A0A182I898 A0A1J1IVC6 A0A026WVJ4 B4GLA6 A0A182N6G1 A0A182YIF0 A0A182VXN3 A0A3L8DRX4 A0A182LY04 A0A336MBF4 A0A151IIR9 A0A195DNA5 A0A182KMQ3 Q7Z1M1 W5J3B1 A0A182XH54 A0A182VKT6 A0A195BNC5 A0A195F008 A0A0L7RD10 A0A0M9AB20 A0A151WYM6 F4X5H3 A0A034WKT0 A0A158P2Y4 A0A1B0FPG8 A0A232FBP7 A0A1B0CI43 A0A182QEB7 E2C3G1 A0A310SJ10 K7J3D0 A0A154P1X9 A0A088AMG9 A0A2A3EHF4 E0VUA6 Q1L729 A0A2R7W7D1 B0X562 A0A1I8JUR3 T1IDW3 N6U917 A0A0J7KM20 A0A1J1HFB9 E2AUK2 A0A1D2N676 A0A2P8YM68 K1RSL2 A0A226EKZ0 A0A0K2UPY8 A0A1S3IHP3
B4N8M1 A0A0M3QYI4 A0A1S4FGF9 A0A1I8P675 Q9VFA9 B4LZZ8 B4HDZ9 B4JGC0 Q294C0 A0A1W4VMK1 A0A1A9ZY94 A0A1A9XWD0 A0A3B0KCL2 A0A034WGQ2 W8BZ14 A0A1B0BL43 A0A3B0KJK5 A0A1A9VAX2 B3MX93 B4PS70 A0A0R1E419 B3P0S7 A0A067QRF1 A0A0L0BPN5 A0A2J7QL05 A0A084W5B9 A0A182ITC2 A0A139WEF3 A0A182PF20 A0A182F835 A0A139WE35 A0A1Y1KLR8 A0A336MI61 A0A336MBT3 A0A182I898 A0A1J1IVC6 A0A026WVJ4 B4GLA6 A0A182N6G1 A0A182YIF0 A0A182VXN3 A0A3L8DRX4 A0A182LY04 A0A336MBF4 A0A151IIR9 A0A195DNA5 A0A182KMQ3 Q7Z1M1 W5J3B1 A0A182XH54 A0A182VKT6 A0A195BNC5 A0A195F008 A0A0L7RD10 A0A0M9AB20 A0A151WYM6 F4X5H3 A0A034WKT0 A0A158P2Y4 A0A1B0FPG8 A0A232FBP7 A0A1B0CI43 A0A182QEB7 E2C3G1 A0A310SJ10 K7J3D0 A0A154P1X9 A0A088AMG9 A0A2A3EHF4 E0VUA6 Q1L729 A0A2R7W7D1 B0X562 A0A1I8JUR3 T1IDW3 N6U917 A0A0J7KM20 A0A1J1HFB9 E2AUK2 A0A1D2N676 A0A2P8YM68 K1RSL2 A0A226EKZ0 A0A0K2UPY8 A0A1S3IHP3
Pubmed
19121390
26354079
17994087
17510324
10731132
12537568
+ More
12537572 12537573 12537574 16110336 17569856 17569867 15632085 23185243 25348373 24495485 17550304 24845553 26108605 24438588 18362917 19820115 28004739 24508170 25244985 30249741 20966253 12364791 14747013 17210077 20920257 23761445 21719571 21347285 28648823 20798317 20075255 20566863 23537049 27289101 29403074 22992520
12537572 12537573 12537574 16110336 17569856 17569867 15632085 23185243 25348373 24495485 17550304 24845553 26108605 24438588 18362917 19820115 28004739 24508170 25244985 30249741 20966253 12364791 14747013 17210077 20920257 23761445 21719571 21347285 28648823 20798317 20075255 20566863 23537049 27289101 29403074 22992520
EMBL
BABH01017390
BABH01017391
BABH01017392
KQ460890
KPJ10885.1
KQ459593
+ More
KPI96476.1 NWSH01005380 PCG64258.1 ODYU01003411 SOQ42145.1 CH933806 EDW15171.1 CH964232 EDW81472.1 CP012526 ALC47733.1 AE014297 AAF55151.2 CH940650 EDW67226.1 CH480815 EDW42091.1 CH916369 EDV92589.1 CM000070 EAL29044.2 OUUW01000008 SPP83939.1 GAKP01004226 JAC54726.1 GAMC01002003 JAC04553.1 JXJN01016211 SPP83938.1 CH902629 EDV35316.1 CM000160 EDW97490.2 KRK03775.1 CH954181 EDV48975.1 KK853050 KDR12066.1 JRES01001565 KNC22050.1 NEVH01013255 PNF29238.1 ATLV01020573 ATLV01020574 ATLV01020575 KE525303 KFB45413.1 KQ971354 KYB26237.1 KYB26238.1 GEZM01086085 JAV59787.1 UFQT01000610 SSX25708.1 UFQT01000824 SSX27410.1 APCN01002684 CVRI01000063 CRL04219.1 KK107099 EZA59681.1 CH479185 EDW38330.1 QOIP01000005 RLU23151.1 AXCM01004698 SSX25707.1 KQ977459 KYN02566.1 KQ980713 KYN14385.1 AF510719 AAAB01008978 AAP47148.1 EAA13613.3 ADMH02002112 ETN58837.1 KQ976432 KYM87530.1 KQ981905 KYN33459.1 KQ414615 KOC68680.1 KQ435711 KOX79613.1 KQ982649 KYQ52979.1 GL888716 EGI58329.1 GAKP01004227 JAC54725.1 ADTU01007645 CCAG010007785 NNAY01000520 OXU27883.1 AJWK01013033 AXCN02000286 GL452320 EFN77499.1 KQ762882 OAD55372.1 KQ434796 KZC05832.1 KZ288248 PBC31140.1 DS235784 EEB16962.1 DQ011227 AAY63896.1 KK854411 PTY15622.1 DS232366 EDS40708.1 ACPB03002174 APGK01037738 KB740949 ENN77146.1 LBMM01005645 KMQ91337.1 CVRI01000002 CRK86663.1 GL442829 EFN62896.1 LJIJ01000187 ODN00770.1 PYGN01000498 PSN45323.1 JH818095 EKC37511.1 LNIX01000003 OXA58363.1 HACA01022445 CDW39806.1
KPI96476.1 NWSH01005380 PCG64258.1 ODYU01003411 SOQ42145.1 CH933806 EDW15171.1 CH964232 EDW81472.1 CP012526 ALC47733.1 AE014297 AAF55151.2 CH940650 EDW67226.1 CH480815 EDW42091.1 CH916369 EDV92589.1 CM000070 EAL29044.2 OUUW01000008 SPP83939.1 GAKP01004226 JAC54726.1 GAMC01002003 JAC04553.1 JXJN01016211 SPP83938.1 CH902629 EDV35316.1 CM000160 EDW97490.2 KRK03775.1 CH954181 EDV48975.1 KK853050 KDR12066.1 JRES01001565 KNC22050.1 NEVH01013255 PNF29238.1 ATLV01020573 ATLV01020574 ATLV01020575 KE525303 KFB45413.1 KQ971354 KYB26237.1 KYB26238.1 GEZM01086085 JAV59787.1 UFQT01000610 SSX25708.1 UFQT01000824 SSX27410.1 APCN01002684 CVRI01000063 CRL04219.1 KK107099 EZA59681.1 CH479185 EDW38330.1 QOIP01000005 RLU23151.1 AXCM01004698 SSX25707.1 KQ977459 KYN02566.1 KQ980713 KYN14385.1 AF510719 AAAB01008978 AAP47148.1 EAA13613.3 ADMH02002112 ETN58837.1 KQ976432 KYM87530.1 KQ981905 KYN33459.1 KQ414615 KOC68680.1 KQ435711 KOX79613.1 KQ982649 KYQ52979.1 GL888716 EGI58329.1 GAKP01004227 JAC54725.1 ADTU01007645 CCAG010007785 NNAY01000520 OXU27883.1 AJWK01013033 AXCN02000286 GL452320 EFN77499.1 KQ762882 OAD55372.1 KQ434796 KZC05832.1 KZ288248 PBC31140.1 DS235784 EEB16962.1 DQ011227 AAY63896.1 KK854411 PTY15622.1 DS232366 EDS40708.1 ACPB03002174 APGK01037738 KB740949 ENN77146.1 LBMM01005645 KMQ91337.1 CVRI01000002 CRK86663.1 GL442829 EFN62896.1 LJIJ01000187 ODN00770.1 PYGN01000498 PSN45323.1 JH818095 EKC37511.1 LNIX01000003 OXA58363.1 HACA01022445 CDW39806.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000009192
UP000007798
+ More
UP000092553 UP000095300 UP000000803 UP000008792 UP000001292 UP000001070 UP000001819 UP000192221 UP000092445 UP000092443 UP000268350 UP000092460 UP000078200 UP000007801 UP000002282 UP000008711 UP000027135 UP000037069 UP000235965 UP000030765 UP000075880 UP000007266 UP000075885 UP000069272 UP000075840 UP000183832 UP000053097 UP000008744 UP000075884 UP000076408 UP000075920 UP000279307 UP000075883 UP000078542 UP000078492 UP000075882 UP000007062 UP000000673 UP000076407 UP000075903 UP000078540 UP000078541 UP000053825 UP000053105 UP000075809 UP000007755 UP000005205 UP000092444 UP000215335 UP000092461 UP000075886 UP000008237 UP000002358 UP000076502 UP000005203 UP000242457 UP000009046 UP000002320 UP000075900 UP000015103 UP000019118 UP000036403 UP000000311 UP000094527 UP000245037 UP000005408 UP000198287 UP000085678
UP000092553 UP000095300 UP000000803 UP000008792 UP000001292 UP000001070 UP000001819 UP000192221 UP000092445 UP000092443 UP000268350 UP000092460 UP000078200 UP000007801 UP000002282 UP000008711 UP000027135 UP000037069 UP000235965 UP000030765 UP000075880 UP000007266 UP000075885 UP000069272 UP000075840 UP000183832 UP000053097 UP000008744 UP000075884 UP000076408 UP000075920 UP000279307 UP000075883 UP000078542 UP000078492 UP000075882 UP000007062 UP000000673 UP000076407 UP000075903 UP000078540 UP000078541 UP000053825 UP000053105 UP000075809 UP000007755 UP000005205 UP000092444 UP000215335 UP000092461 UP000075886 UP000008237 UP000002358 UP000076502 UP000005203 UP000242457 UP000009046 UP000002320 UP000075900 UP000015103 UP000019118 UP000036403 UP000000311 UP000094527 UP000245037 UP000005408 UP000198287 UP000085678
Pfam
Interpro
IPR029020
Ammonium/urea_transptr
+ More
IPR024041 NH4_transpt_AmtB-like_dom
IPR001905 Ammonium_transpt
IPR005821 Ion_trans_dom
IPR006153 Cation/H_exchanger
IPR014710 RmlC-like_jellyroll
IPR018422 Cation/H_exchanger_CPA1
IPR027359 Volt_channel_dom_sf
IPR000595 cNMP-bd_dom
IPR018490 cNMP-bd-like
IPR012934 Znf_AD
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR004241 Atg8-like
IPR029071 Ubiquitin-like_domsf
IPR024041 NH4_transpt_AmtB-like_dom
IPR001905 Ammonium_transpt
IPR005821 Ion_trans_dom
IPR006153 Cation/H_exchanger
IPR014710 RmlC-like_jellyroll
IPR018422 Cation/H_exchanger_CPA1
IPR027359 Volt_channel_dom_sf
IPR000595 cNMP-bd_dom
IPR018490 cNMP-bd-like
IPR012934 Znf_AD
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR004241 Atg8-like
IPR029071 Ubiquitin-like_domsf
Gene 3D
CDD
ProteinModelPortal
H9JJM3
A0A194R0V9
A0A194PSR8
A0A2A4IXX3
A0A2H1VMS8
B4K5V9
+ More
B4N8M1 A0A0M3QYI4 A0A1S4FGF9 A0A1I8P675 Q9VFA9 B4LZZ8 B4HDZ9 B4JGC0 Q294C0 A0A1W4VMK1 A0A1A9ZY94 A0A1A9XWD0 A0A3B0KCL2 A0A034WGQ2 W8BZ14 A0A1B0BL43 A0A3B0KJK5 A0A1A9VAX2 B3MX93 B4PS70 A0A0R1E419 B3P0S7 A0A067QRF1 A0A0L0BPN5 A0A2J7QL05 A0A084W5B9 A0A182ITC2 A0A139WEF3 A0A182PF20 A0A182F835 A0A139WE35 A0A1Y1KLR8 A0A336MI61 A0A336MBT3 A0A182I898 A0A1J1IVC6 A0A026WVJ4 B4GLA6 A0A182N6G1 A0A182YIF0 A0A182VXN3 A0A3L8DRX4 A0A182LY04 A0A336MBF4 A0A151IIR9 A0A195DNA5 A0A182KMQ3 Q7Z1M1 W5J3B1 A0A182XH54 A0A182VKT6 A0A195BNC5 A0A195F008 A0A0L7RD10 A0A0M9AB20 A0A151WYM6 F4X5H3 A0A034WKT0 A0A158P2Y4 A0A1B0FPG8 A0A232FBP7 A0A1B0CI43 A0A182QEB7 E2C3G1 A0A310SJ10 K7J3D0 A0A154P1X9 A0A088AMG9 A0A2A3EHF4 E0VUA6 Q1L729 A0A2R7W7D1 B0X562 A0A1I8JUR3 T1IDW3 N6U917 A0A0J7KM20 A0A1J1HFB9 E2AUK2 A0A1D2N676 A0A2P8YM68 K1RSL2 A0A226EKZ0 A0A0K2UPY8 A0A1S3IHP3
B4N8M1 A0A0M3QYI4 A0A1S4FGF9 A0A1I8P675 Q9VFA9 B4LZZ8 B4HDZ9 B4JGC0 Q294C0 A0A1W4VMK1 A0A1A9ZY94 A0A1A9XWD0 A0A3B0KCL2 A0A034WGQ2 W8BZ14 A0A1B0BL43 A0A3B0KJK5 A0A1A9VAX2 B3MX93 B4PS70 A0A0R1E419 B3P0S7 A0A067QRF1 A0A0L0BPN5 A0A2J7QL05 A0A084W5B9 A0A182ITC2 A0A139WEF3 A0A182PF20 A0A182F835 A0A139WE35 A0A1Y1KLR8 A0A336MI61 A0A336MBT3 A0A182I898 A0A1J1IVC6 A0A026WVJ4 B4GLA6 A0A182N6G1 A0A182YIF0 A0A182VXN3 A0A3L8DRX4 A0A182LY04 A0A336MBF4 A0A151IIR9 A0A195DNA5 A0A182KMQ3 Q7Z1M1 W5J3B1 A0A182XH54 A0A182VKT6 A0A195BNC5 A0A195F008 A0A0L7RD10 A0A0M9AB20 A0A151WYM6 F4X5H3 A0A034WKT0 A0A158P2Y4 A0A1B0FPG8 A0A232FBP7 A0A1B0CI43 A0A182QEB7 E2C3G1 A0A310SJ10 K7J3D0 A0A154P1X9 A0A088AMG9 A0A2A3EHF4 E0VUA6 Q1L729 A0A2R7W7D1 B0X562 A0A1I8JUR3 T1IDW3 N6U917 A0A0J7KM20 A0A1J1HFB9 E2AUK2 A0A1D2N676 A0A2P8YM68 K1RSL2 A0A226EKZ0 A0A0K2UPY8 A0A1S3IHP3
PDB
5FUF
E-value=6.67444e-40,
Score=414
Ontologies
GO
PANTHER
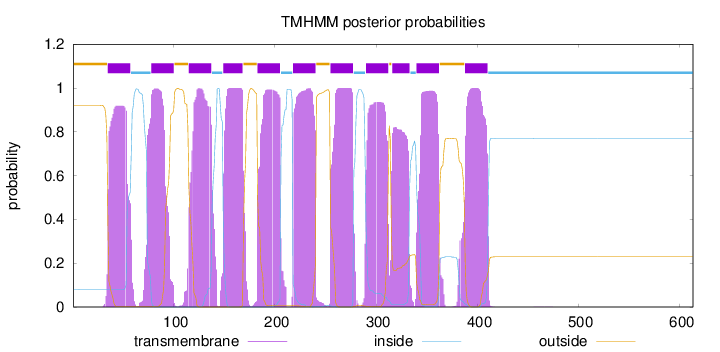
Topology
Subcellular location
Membrane
Length:
613
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
231.03274
Exp number, first 60 AAs:
19.39782
Total prob of N-in:
0.07913
POSSIBLE N-term signal
sequence
outside
1 - 34
TMhelix
35 - 57
inside
58 - 77
TMhelix
78 - 100
outside
101 - 114
TMhelix
115 - 137
inside
138 - 148
TMhelix
149 - 168
outside
169 - 182
TMhelix
183 - 205
inside
206 - 217
TMhelix
218 - 240
outside
241 - 254
TMhelix
255 - 277
inside
278 - 289
TMhelix
290 - 312
outside
313 - 315
TMhelix
316 - 333
inside
334 - 339
TMhelix
340 - 362
outside
363 - 387
TMhelix
388 - 410
inside
411 - 613
Population Genetic Test Statistics
Pi
26.133447
Theta
174.927866
Tajima's D
2.373298
CLR
0.158273
CSRT
0.928553572321384
Interpretation
Uncertain
