Gene
KWMTBOMO00769
Annotation
non-LTR_retrotransposon_R1Bmks_ORF1_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 0.919
Sequence
CDS
ATGTCGGAGGAGGAGAGGGGGCTATTTTCCCCTCGGGTATCTTTGGCGCGCTCGCCGCCTAGGGGACTGACTGCACCTCTGCCGGCGCCTATGCCGCCGCCGGCCCCCCGGGGGGTTAGGGCGGGAGCGGCTGTGAGTGCCAAAGGCCGGAGGACCGGCCTGCATATCCCGGTTGCTTCTGGACTGTCGTCCTCGGCGCCTGTCACCCCGGTTGACCCAGAGGCTGGTGTCCCTAGCTTCCCCATTCCTGTTGCGTCGGGGGCTCCGACTGGACCCCTTGATGTAGCAGCGCAGGGGAGGCTGGAGCTGCTCGAGCGGGCAAACAGAGCGGTGCGCGGGATCATGTCGGTGGCGACGGCTGCATCTAAGCTCAATAAGTCGGAGGTCAATCTAATCTCCGAACTCGGTCGCGATATCCTGGCCGTAGTTGGGGCCCTCGGGATCCAGCTCTCTGATAAGGAGCTGGTGGTCGAAAGGCTCCGCTCGGCAGAGGCCTCTGCACGGCGTGACTCCGTGGCGCCTTCGATGGCTGCTGGTGCGGCGGCCGGGTCTGGGACTGGGCCTGCGACCTTCGCCACTGTCCTCAGGACGGGCCCTGGTGGGGTCCCGAGGTCCATTGGGGCCTCTCAGGGGCCTTCCTTGGCATTCTACCCGTCCGAGGGCAATGCGGAACTGAAGACAGCTGAGGACACAAAGAAAGAGGTAAAAAAGGCCATTGACCCTAAGTCAATGGCTATTGGAATACAGAGCGTAAGGAAGGTTGGGAATGCGGGGGTAGTGGTGCAGACCACGTCCCCCGCGGCCGCAGTCAAGCTTAGAAATGCTGCCCCGCCATCACTGAGAGTCACTGAACCCAGGCGCCGACAGCCCTTAGTAGCTGTCAATGGCGTGGAGGGCGACCCCTCCTTCGAGGAGGTAATCGAGTGCCTGGCTAGCCAGAACCTCGACCCGGAGGAGTGGCCCCTCACTAGGGTACGAGCGGAGCTCACGGGGGCGTTCAAAAAGGGGAGGCGACAATCCAACAACACTACTGTGGTGTTTAACGCCTCCCCTCGCATCAGGGACGCCCTCGTGAAGATTGGCAGAGTGTATGTGGGGTGGGTCGCCTGTGAGGTCACGGACTTTGTCCGGGTGACCTGCTGCAATAAATGTCAGCAATATGGTCACCCGGAGAAATTTTGCCGGGCCAAGGAGGCCACCTGTGGCCGATGTGGAGAGGATGGCCACCGAATGGAGGCCTTGCACATTGGGGGGTGGGACTTATATATGGTGTCTGCGTACTTCCAGTATAGTGACCCTATTGACCCATACCTGCACCGGCTCGGGAATATTCTTGACCGGCTGCGGGGGGCTCGGGTCGTTATCTGCGCAGACACTAATGCCCACTCGCCATTGTGGCACTCGCTGCCCAGGCACTACGTCGGTCGGGGTCAGGAAGTGGCTGACCGCCGCGCCAAGATGGAGGATTTCATTGGGGCGAGGCGGTTGGTCGTCCATAACGCGGATGGCCACCTGCCGACCTTCAGTACGGCGAACGGAGAATCTTATGTCGATGTCACGCTGTCTACGCGGGGAGTACGCGTGTCTGAATGGCGTGTAACTAATGAATCATCGAGCGATCACCGGCTCATTGTGTTTGGGGTGGGGGGCGATACAACAGGGGAGCGGAACGAGGACGAGGAGGCGCGGAGCGATTTGAGGCCGGATGAGCCGTGCGCACTGCGTCGGTACCGGGACCGTGGGGTGGATTGGGACCTCTTCAGATCGCGTATCCATGAGCGAATGGGGAGTCTGGACCTCGAGGAACCTGTGGCTGCCCTTTGCGAAAAATTTACCGGGGTTATAACTCGCACCGCTGCAGAATGCTTAGGATCACTGAAAGCAGATAGAACTGACAGGGGTTATGAATGGTGGACCCCAGTACTCGATAAGCTTAGGGTAGCCCAGGGTAGGGCCAGGCGTCGATGGCAAAAGGCCCGCCGAACAGGGGGTGAGGAAGAGGAGCAGTCTGGGAGAGTCTTCCGCGACCGCAGGCGCGAGTATCGAAGAGCGATGCATGACGCTGAGACCGCCTTTTACCGGGAGATCGCTGAAGGGGGAAACCATGACCCGTGGGGACTAGCGTATCGGACGGCGAGTGGTAGGCGACGCGCACCAACTAATGTGGTTAACGGGGTGGAGTATGCGGGGCGGTGCTCGGATGATGTGAGTGGGGCCATGCGCACCTTGATGTGGGCGCTGTGTCCGGATGACTATATGTCTCGGGACACTCCGTACCATGCGCGGGTGCGTATCATGGCCGCGCTCCCCCCATCCGGGCGGGACGCAGACCCGCTGAGCAAAGACTCCCTTCGTGCCATAATTGGCTCACTGAAGAATACCGCACCGGGCATCGACGGTTTAACGGCGCGCATTATCAAAAAGGCACTTCCGGCTGCTGAGGCCGAGTTCGTGGCCGTATACGCACGCACGGCTGCTTTGCCGGTGTTGGCGGGCGTCCTGCCGGCGGACCTGGAGGTGACTCGTGCTGGACGGAGGATGCGGGAGTGCGAAGGATTGGCGCGGGAGTTGGCGGCGGAGAGACGACGGCGGATCGACGGCGATGTCTTGGTAGTTTGGCAGGACAGGTGGGCGTCTGAGGGTAAGGGGAGGGAACTGTACAAGTTCTTTCCCGATGTTGCGGACAGGAAGAAGGCAACGTGGATGGAGCCGGACTATCAGACCTCGCAGATCCTCACGGGACATGGGATCTTCAATAAGCGATTGGCGGATATGCGACTGAGGGAGGGGCATGCTTGCGACTGCGGGGCGGTTGAGGAGGACAGGGACCATGTCCTGTGGGAGTGTCCTCTCTATGACGAAATCCGGGGCAGGATGCTCGATGGAATCTCGCGGTCTGAGGTGGGCCCAGTTTACCACGCGGACCTGGTCAGGGACGAGAAAAATTTTCGGCTCTTGCGCGAGTTCGCGCATACATGGCACACGGCGCGCACTGCGCGCGAATTACGAGGACTGCCTGGCGACGAAGAATGTGGGTGA
Protein
MSEEERGLFSPRVSLARSPPRGLTAPLPAPMPPPAPRGVRAGAAVSAKGRRTGLHIPVASGLSSSAPVTPVDPEAGVPSFPIPVASGAPTGPLDVAAQGRLELLERANRAVRGIMSVATAASKLNKSEVNLISELGRDILAVVGALGIQLSDKELVVERLRSAEASARRDSVAPSMAAGAAAGSGTGPATFATVLRTGPGGVPRSIGASQGPSLAFYPSEGNAELKTAEDTKKEVKKAIDPKSMAIGIQSVRKVGNAGVVVQTTSPAAAVKLRNAAPPSLRVTEPRRRQPLVAVNGVEGDPSFEEVIECLASQNLDPEEWPLTRVRAELTGAFKKGRRQSNNTTVVFNASPRIRDALVKIGRVYVGWVACEVTDFVRVTCCNKCQQYGHPEKFCRAKEATCGRCGEDGHRMEALHIGGWDLYMVSAYFQYSDPIDPYLHRLGNILDRLRGARVVICADTNAHSPLWHSLPRHYVGRGQEVADRRAKMEDFIGARRLVVHNADGHLPTFSTANGESYVDVTLSTRGVRVSEWRVTNESSSDHRLIVFGVGGDTTGERNEDEEARSDLRPDEPCALRRYRDRGVDWDLFRSRIHERMGSLDLEEPVAALCEKFTGVITRTAAECLGSLKADRTDRGYEWWTPVLDKLRVAQGRARRRWQKARRTGGEEEEQSGRVFRDRRREYRRAMHDAETAFYREIAEGGNHDPWGLAYRTASGRRRAPTNVVNGVEYAGRCSDDVSGAMRTLMWALCPDDYMSRDTPYHARVRIMAALPPSGRDADPLSKDSLRAIIGSLKNTAPGIDGLTARIIKKALPAAEAEFVAVYARTAALPVLAGVLPADLEVTRAGRRMRECEGLARELAAERRRRIDGDVLVVWQDRWASEGKGRELYKFFPDVADRKKATWMEPDYQTSQILTGHGIFNKRLADMRLREGHACDCGAVEEDRDHVLWECPLYDEIRGRMLDGISRSEVGPVYHADLVRDEKNFRLLREFAHTWHTARTARELRGLPGDEECG
Summary
Uniprot
ProteinModelPortal
PDB
2EI9
E-value=6.0342e-81,
Score=770
Ontologies
GO
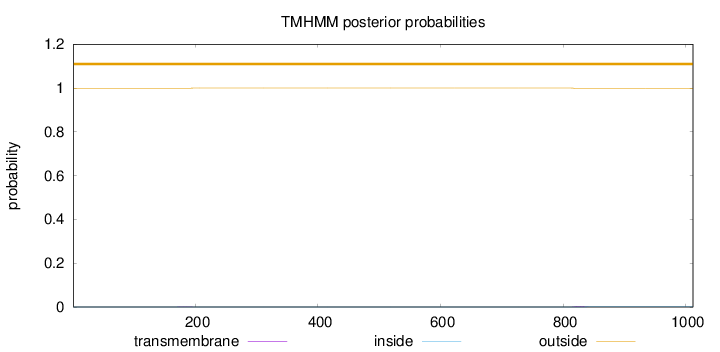
Topology
Length:
1012
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03967
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.00049
outside
1 - 1012
Population Genetic Test Statistics
Pi
1.137605
Theta
0.968781
Tajima's D
0
CLR
0.800234
CSRT
0.434578271086446
Interpretation
Uncertain
