Gene
KWMTBOMO00751
Pre Gene Modal
BGIBMGA013339
Annotation
ornithine_decarboxylase_antizyme_[Bombyx_mori]
Full name
Ornithine decarboxylase antizyme 1
+ More
Ornithine decarboxylase antizyme
Ornithine decarboxylase antizyme
Alternative Name
ODC antizyme, short form
Location in the cell
Extracellular Reliability : 1.481 Nuclear Reliability : 1.193
Sequence
CDS
ATGATGTACCTCCGCGTGCCCGGGGTCCTGCAGTCCGGCAGCAAGGACAGCTTCATGCTGCTCCTGGATTTCGCTGAAGAGCGTCTCGGCTGTAAGAGTTGCATCATCTGCGTTTTGAAGAGTCGTCCCGACCGCGCTACCCTGCTGCGCACCTTCATGTTCATGGGATTCCAAGTTCTGGCTCCGAATTCGCCGCTGACCCCACAGCACATCAACAACCCTAATTACATTTTCTTACACTACAATATGCAGTAA
Protein
MMYLRVPGVLQSGSKDSFMLLLDFAEERLGCKSCIICVLKSRPDRATLLRTFMFMGFQVLAPNSPLTPQHINNPNYIFLHYNMQ
Summary
Description
Ornithine decarboxylase (ODC) antizyme protein that negatively regulates ODC activity and intracellular polyamine biosynthesis and uptake in response to increased intracellular polyamine levels. Binds to ODC monomers, inhibiting the assembly of the functional ODC homodimer, and targets the monomers for ubiquitin-independent proteolytic destruction by the 26S proteasome.
Subunit
Interacts with ODC1 and thereby sterically blocks ODC homodimerization.
Similarity
Belongs to the CD36 family.
Belongs to the ODC antizyme family.
Belongs to the ODC antizyme family.
Keywords
Complete proteome
Polyamine biosynthesis
Reference proteome
Ribosomal frameshifting
Feature
chain Ornithine decarboxylase antizyme 1
Uniprot
H9JUX6
Q9GSV9
A0A0K2S3N3
A0A3B0EV17
A0A194PU31
A0A2H1V4P6
+ More
A0A2W1BGI7 A0A194R4U9 A0A2A4K061 A0A2A4K0I7 A0A2A4JZ44 A0A0K8S867 A0A212EXT8 A0A2Z6FLD0 A0A1J1HNH0 A0A1J1HSZ5 C3ZX52 A0A1L8E2N1 A0A1B0D1T9 A0A1S4H3Y5 A0A182HQX2 A0A182WXJ2 A0A3N0Y3N8 A0A1B0CKD8 A0A023END3 A0A182FN08 Q9YI98 A0A384BH35 A0A182IYX0 A0A1S4H3U0 A0A2U9C3B0 A0A182VDG5 A0A084VWW2 A0A182KJD5 R7VI46 L5MIF9 A0A1A8ILR5 A0A1A7ZE21 A0A1A8P1A4 A0A1A8LJH8 A0A1A8BPB1 A0A1A8HF04 A0A182UK52 A0A2M3YXK9 A0A2M4A0U7 Q95P51 A0A182K677 A0A3B5AME1 L5ME86 A0A182T4N3 A0A023EMB1 A0A1A8VF33 A0A1A7X5P3 A0A182Q4G2 A0A182YQV2 A0A182MIK8 A0A1A8QA68 A7RKX2 A0A2M4CKR8 A0A1W7R8J5 H0YPX3 A0A3B5AL80 A0A3M0JJ38 W5JQA2 A0A3B4GGH8 A0A3P8QW86 A0A3P9BF37 T1DRW6 A0A146NQN9 A0A315VNU8 Q6X3F2 S7MG55 A0A182PTL5 A0A2P4SQL1 A5JST0 I3MZW7 A0A3B3X8E6 A0A2G9Q4H0 I3JAY0 Q5TNW3 A0A0Q3QYA8 A0A096MH89 K1RDR6 A0A3B3W2J2 L5L6I4 A0A2V2PAT3 F6VR40 A0A1Y9GH65 A0A3B4YLY6 A0A3B4TML7 A0A2I4AX82 A0A3P8SAR7 M7AKR9 A0A182NNN8 A0A060XBG3 A0A3B3X8B0 A0A226M745 A0A151NGV5 A0A182W8E2 A0A3B4Y7C9
A0A2W1BGI7 A0A194R4U9 A0A2A4K061 A0A2A4K0I7 A0A2A4JZ44 A0A0K8S867 A0A212EXT8 A0A2Z6FLD0 A0A1J1HNH0 A0A1J1HSZ5 C3ZX52 A0A1L8E2N1 A0A1B0D1T9 A0A1S4H3Y5 A0A182HQX2 A0A182WXJ2 A0A3N0Y3N8 A0A1B0CKD8 A0A023END3 A0A182FN08 Q9YI98 A0A384BH35 A0A182IYX0 A0A1S4H3U0 A0A2U9C3B0 A0A182VDG5 A0A084VWW2 A0A182KJD5 R7VI46 L5MIF9 A0A1A8ILR5 A0A1A7ZE21 A0A1A8P1A4 A0A1A8LJH8 A0A1A8BPB1 A0A1A8HF04 A0A182UK52 A0A2M3YXK9 A0A2M4A0U7 Q95P51 A0A182K677 A0A3B5AME1 L5ME86 A0A182T4N3 A0A023EMB1 A0A1A8VF33 A0A1A7X5P3 A0A182Q4G2 A0A182YQV2 A0A182MIK8 A0A1A8QA68 A7RKX2 A0A2M4CKR8 A0A1W7R8J5 H0YPX3 A0A3B5AL80 A0A3M0JJ38 W5JQA2 A0A3B4GGH8 A0A3P8QW86 A0A3P9BF37 T1DRW6 A0A146NQN9 A0A315VNU8 Q6X3F2 S7MG55 A0A182PTL5 A0A2P4SQL1 A5JST0 I3MZW7 A0A3B3X8E6 A0A2G9Q4H0 I3JAY0 Q5TNW3 A0A0Q3QYA8 A0A096MH89 K1RDR6 A0A3B3W2J2 L5L6I4 A0A2V2PAT3 F6VR40 A0A1Y9GH65 A0A3B4YLY6 A0A3B4TML7 A0A2I4AX82 A0A3P8SAR7 M7AKR9 A0A182NNN8 A0A060XBG3 A0A3B3X8B0 A0A226M745 A0A151NGV5 A0A182W8E2 A0A3B4Y7C9
Pubmed
EMBL
BABH01041439
BABH01041440
AF291575
AAG16235.1
AB999997
BAS21455.1
+ More
RBVL01000177 RKO11078.1 KQ459593 KPI96483.1 ODYU01000657 SOQ35811.1 KZ150197 PZC72317.1 KQ460890 KPJ10891.1 NWSH01000338 PCG77284.1 PCG77283.1 PCG77285.1 GBRD01016353 JAG49473.1 AGBW02011708 OWR46254.1 LC384840 BBE28983.1 CVRI01000013 CRK89599.1 CRK89598.1 GG666708 EEN42881.1 GFDF01001107 JAV12977.1 AJVK01010393 AJVK01010394 AAAB01008980 APCN01001608 RJVU01053127 ROL40797.1 AJWK01016088 AJWK01016089 AJWK01016090 GAPW01003183 JAC10415.1 AB017117 CP026254 AWP11104.1 ATLV01017759 KE525185 KFB42456.1 AMQN01003837 KB292015 ELU18279.1 KB100339 ELK37503.1 HAED01011810 SBQ98204.1 HADY01002582 SBP41067.1 HAEI01001181 SBR75043.1 HAEF01007104 HAEG01007493 SBR44486.1 HADZ01004752 HAEA01013847 SBP68693.1 HAEB01008266 HAEC01013048 SBQ81265.1 GGFM01000249 MBW21000.1 GGFK01001082 MBW34403.1 AF396870 AF396871 AY064120 AY133346 CH477314 KB101390 ELK36647.1 GAPW01004154 JAC09444.1 HAEJ01018024 SBS58481.1 HADW01011835 HADX01005021 SBP13235.1 AXCN02001967 AXCM01005077 HAEH01010580 SBR90079.1 DS469517 EDO47835.1 GGFL01001657 MBW65835.1 GEHC01000164 JAV47481.1 ABQF01047514 QRBI01000142 RMC00829.1 ADMH02000643 ETN65468.1 GAMD01000460 JAB01131.1 GCES01152570 JAQ33752.1 NHOQ01001926 PWA20921.1 AY257551 AAP82032.1 KE161324 EPQ03134.1 PPHD01028928 POI26404.1 EF564268 ABQ51195.1 AGTP01068775 AGTP01068776 KZ369196 PIO10445.1 AERX01026419 EAL39277.1 LMAW01002688 KQK78097.1 AYCK01018215 JH818863 EKC32251.1 KB030265 ELK19339.1 QGNU01000250 PWS23116.1 KB586660 EMP25741.1 FR905190 CDQ76978.1 AWGT02031212 OXB51070.1 AKHW03003034 KYO36018.1
RBVL01000177 RKO11078.1 KQ459593 KPI96483.1 ODYU01000657 SOQ35811.1 KZ150197 PZC72317.1 KQ460890 KPJ10891.1 NWSH01000338 PCG77284.1 PCG77283.1 PCG77285.1 GBRD01016353 JAG49473.1 AGBW02011708 OWR46254.1 LC384840 BBE28983.1 CVRI01000013 CRK89599.1 CRK89598.1 GG666708 EEN42881.1 GFDF01001107 JAV12977.1 AJVK01010393 AJVK01010394 AAAB01008980 APCN01001608 RJVU01053127 ROL40797.1 AJWK01016088 AJWK01016089 AJWK01016090 GAPW01003183 JAC10415.1 AB017117 CP026254 AWP11104.1 ATLV01017759 KE525185 KFB42456.1 AMQN01003837 KB292015 ELU18279.1 KB100339 ELK37503.1 HAED01011810 SBQ98204.1 HADY01002582 SBP41067.1 HAEI01001181 SBR75043.1 HAEF01007104 HAEG01007493 SBR44486.1 HADZ01004752 HAEA01013847 SBP68693.1 HAEB01008266 HAEC01013048 SBQ81265.1 GGFM01000249 MBW21000.1 GGFK01001082 MBW34403.1 AF396870 AF396871 AY064120 AY133346 CH477314 KB101390 ELK36647.1 GAPW01004154 JAC09444.1 HAEJ01018024 SBS58481.1 HADW01011835 HADX01005021 SBP13235.1 AXCN02001967 AXCM01005077 HAEH01010580 SBR90079.1 DS469517 EDO47835.1 GGFL01001657 MBW65835.1 GEHC01000164 JAV47481.1 ABQF01047514 QRBI01000142 RMC00829.1 ADMH02000643 ETN65468.1 GAMD01000460 JAB01131.1 GCES01152570 JAQ33752.1 NHOQ01001926 PWA20921.1 AY257551 AAP82032.1 KE161324 EPQ03134.1 PPHD01028928 POI26404.1 EF564268 ABQ51195.1 AGTP01068775 AGTP01068776 KZ369196 PIO10445.1 AERX01026419 EAL39277.1 LMAW01002688 KQK78097.1 AYCK01018215 JH818863 EKC32251.1 KB030265 ELK19339.1 QGNU01000250 PWS23116.1 KB586660 EMP25741.1 FR905190 CDQ76978.1 AWGT02031212 OXB51070.1 AKHW03003034 KYO36018.1
Proteomes
UP000005204
UP000276569
UP000053268
UP000053240
UP000218220
UP000007151
+ More
UP000183832 UP000001554 UP000092462 UP000075840 UP000076407 UP000092461 UP000069272 UP000000437 UP000075880 UP000246464 UP000075903 UP000030765 UP000075882 UP000014760 UP000075902 UP000008820 UP000075881 UP000261400 UP000075901 UP000075886 UP000076408 UP000075883 UP000001593 UP000007754 UP000269221 UP000000673 UP000261460 UP000265100 UP000265160 UP000075885 UP000005215 UP000261480 UP000005207 UP000007062 UP000051836 UP000028760 UP000005408 UP000261500 UP000010552 UP000245538 UP000002280 UP000261360 UP000261420 UP000192220 UP000265080 UP000031443 UP000075884 UP000193380 UP000198419 UP000050525 UP000075920
UP000183832 UP000001554 UP000092462 UP000075840 UP000076407 UP000092461 UP000069272 UP000000437 UP000075880 UP000246464 UP000075903 UP000030765 UP000075882 UP000014760 UP000075902 UP000008820 UP000075881 UP000261400 UP000075901 UP000075886 UP000076408 UP000075883 UP000001593 UP000007754 UP000269221 UP000000673 UP000261460 UP000265100 UP000265160 UP000075885 UP000005215 UP000261480 UP000005207 UP000007062 UP000051836 UP000028760 UP000005408 UP000261500 UP000010552 UP000245538 UP000002280 UP000261360 UP000261420 UP000192220 UP000265080 UP000031443 UP000075884 UP000193380 UP000198419 UP000050525 UP000075920
Interpro
Gene 3D
ProteinModelPortal
H9JUX6
Q9GSV9
A0A0K2S3N3
A0A3B0EV17
A0A194PU31
A0A2H1V4P6
+ More
A0A2W1BGI7 A0A194R4U9 A0A2A4K061 A0A2A4K0I7 A0A2A4JZ44 A0A0K8S867 A0A212EXT8 A0A2Z6FLD0 A0A1J1HNH0 A0A1J1HSZ5 C3ZX52 A0A1L8E2N1 A0A1B0D1T9 A0A1S4H3Y5 A0A182HQX2 A0A182WXJ2 A0A3N0Y3N8 A0A1B0CKD8 A0A023END3 A0A182FN08 Q9YI98 A0A384BH35 A0A182IYX0 A0A1S4H3U0 A0A2U9C3B0 A0A182VDG5 A0A084VWW2 A0A182KJD5 R7VI46 L5MIF9 A0A1A8ILR5 A0A1A7ZE21 A0A1A8P1A4 A0A1A8LJH8 A0A1A8BPB1 A0A1A8HF04 A0A182UK52 A0A2M3YXK9 A0A2M4A0U7 Q95P51 A0A182K677 A0A3B5AME1 L5ME86 A0A182T4N3 A0A023EMB1 A0A1A8VF33 A0A1A7X5P3 A0A182Q4G2 A0A182YQV2 A0A182MIK8 A0A1A8QA68 A7RKX2 A0A2M4CKR8 A0A1W7R8J5 H0YPX3 A0A3B5AL80 A0A3M0JJ38 W5JQA2 A0A3B4GGH8 A0A3P8QW86 A0A3P9BF37 T1DRW6 A0A146NQN9 A0A315VNU8 Q6X3F2 S7MG55 A0A182PTL5 A0A2P4SQL1 A5JST0 I3MZW7 A0A3B3X8E6 A0A2G9Q4H0 I3JAY0 Q5TNW3 A0A0Q3QYA8 A0A096MH89 K1RDR6 A0A3B3W2J2 L5L6I4 A0A2V2PAT3 F6VR40 A0A1Y9GH65 A0A3B4YLY6 A0A3B4TML7 A0A2I4AX82 A0A3P8SAR7 M7AKR9 A0A182NNN8 A0A060XBG3 A0A3B3X8B0 A0A226M745 A0A151NGV5 A0A182W8E2 A0A3B4Y7C9
A0A2W1BGI7 A0A194R4U9 A0A2A4K061 A0A2A4K0I7 A0A2A4JZ44 A0A0K8S867 A0A212EXT8 A0A2Z6FLD0 A0A1J1HNH0 A0A1J1HSZ5 C3ZX52 A0A1L8E2N1 A0A1B0D1T9 A0A1S4H3Y5 A0A182HQX2 A0A182WXJ2 A0A3N0Y3N8 A0A1B0CKD8 A0A023END3 A0A182FN08 Q9YI98 A0A384BH35 A0A182IYX0 A0A1S4H3U0 A0A2U9C3B0 A0A182VDG5 A0A084VWW2 A0A182KJD5 R7VI46 L5MIF9 A0A1A8ILR5 A0A1A7ZE21 A0A1A8P1A4 A0A1A8LJH8 A0A1A8BPB1 A0A1A8HF04 A0A182UK52 A0A2M3YXK9 A0A2M4A0U7 Q95P51 A0A182K677 A0A3B5AME1 L5ME86 A0A182T4N3 A0A023EMB1 A0A1A8VF33 A0A1A7X5P3 A0A182Q4G2 A0A182YQV2 A0A182MIK8 A0A1A8QA68 A7RKX2 A0A2M4CKR8 A0A1W7R8J5 H0YPX3 A0A3B5AL80 A0A3M0JJ38 W5JQA2 A0A3B4GGH8 A0A3P8QW86 A0A3P9BF37 T1DRW6 A0A146NQN9 A0A315VNU8 Q6X3F2 S7MG55 A0A182PTL5 A0A2P4SQL1 A5JST0 I3MZW7 A0A3B3X8E6 A0A2G9Q4H0 I3JAY0 Q5TNW3 A0A0Q3QYA8 A0A096MH89 K1RDR6 A0A3B3W2J2 L5L6I4 A0A2V2PAT3 F6VR40 A0A1Y9GH65 A0A3B4YLY6 A0A3B4TML7 A0A2I4AX82 A0A3P8SAR7 M7AKR9 A0A182NNN8 A0A060XBG3 A0A3B3X8B0 A0A226M745 A0A151NGV5 A0A182W8E2 A0A3B4Y7C9
PDB
4ZGY
E-value=3.47862e-16,
Score=200
Ontologies
KEGG
GO
PANTHER
Topology
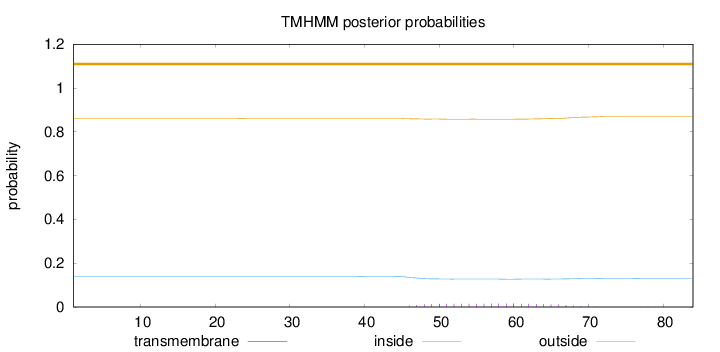
Length:
84
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.30803
Exp number, first 60 AAs:
0.20338
Total prob of N-in:
0.13963
outside
1 - 84
Population Genetic Test Statistics
Pi
186.87404
Theta
146.302445
Tajima's D
1.082416
CLR
0.155133
CSRT
0.678866056697165
Interpretation
Uncertain
