Gene
KWMTBOMO00748
Pre Gene Modal
BGIBMGA013336
Annotation
PREDICTED:_diacylglycerol_kinase_1_isoform_X1_[Papilio_xuthus]
Full name
Diacylglycerol kinase
+ More
Diacylglycerol kinase beta
Diacylglycerol kinase beta
Alternative Name
90 kDa diacylglycerol kinase
Diglyceride kinase beta
Diglyceride kinase beta
Location in the cell
Nuclear Reliability : 2.678
Sequence
CDS
ATGCCTAATCCGGTCGGCGGCGGGTTCCAGTGGGACAAACTCTCGCCGGCCGAGTTTCAACAGCTCCAAGAATTAGCGTCATATTCCACGAGGAAGCTCCAAGATGTGCTCAACGAGTTTTGTGAGGTCAAGAAACAGAGTCCGGACGGGGATATTGACTATGAGGGGTTCCGGTGGTTTCTGGACACGTTCCTCGAGGTCTCCACGCCTGATGAACTCTCTAGACATTTGTTCCTGTCGTTTGTTCGGCGAGATCGCAGACCAGCGCTGCGGGAGATGGCGGCTGCTAGTTCCACTACCGCCTGCGCAGCGGTCACAGCCCATGCTGACCATCAGGTGAAACCAACGCTGGCTTCGAAGATCCACGGTCTGGCTGAGAGACTTCAGCAGCTGGGAAAGAGCTCCGCTGATTCGGTAGACGGTAGCGACAAGGGATCAAGAAGCAGAACCGGTAGCGTCCACCCCATGTTCACGGTGACCACGCATCCTTCGTACTCCTCCCACGAGCTCTGCTTGGACAGACGCAACGAGACCAGTCCCAGTCACAGTCAAATGTCGAGAAACTCCAGCAGAAAGAGCAGCAACTCAATGAGGGTGAACCAGTCTACTAAAATCGAAGACTTTAGACACCTAATGAGACGCTCGTCGACAGTAGAAGTACACACAGCCAGAGTCTCTCTCAAGGATATCGTCTGCTACCTCTCACTCCTAGAAGCAGGCAGACCTGAAGACAAGCTGGAGTTCATGTTTAGACTCTACGACACCGATGGTAACGGCGTACTGGACACTAACGAGATGGACTGCATCGTGAACCAGATGATGGCTGTCGCTGAATATCTAGGCTGGGAGACTTCTGAATTGAGGCCTATTTTACAAGATATGATGGTTGAGATAGACTATGACGCCGATGGGACGGTTTCTCTGGACGAATGGAAACGAGGCGGCATGACCACAATACCTCTGCTGGTTCTCTTGGGCTTGGATACTAACGTGAAAGAAGACGGCGAGCACGCGTGGAGATTAAAGCACTTCAGTAGACCAGCGTATTGCAATCTGTGCTTGAACATGCTCGTGGGGCTCGGGAAAAAGGGCTTGTGTTGCATTTTTTGCAAATTTACGGTACACGAGCGTTGCGTCCAGCGAGCACCGGCCTCTTGCATAGCCACTTATTCCAAGTCCAAGCGTGCTTCTGCCACACTTGCTCATCATTGGATCGAAGGAAATTGCCACGGAAAATGCGCTAGATGTCGGAAAAAGATCAAAGGCTACAACGGTATCACCGGCTTGCACTGTCGCTGGTGTCACATTACACTCCACAACCGTTGCGTGGGCGCGGCGGGCGGGGCGTGCACATTGGGTCGCCACGCGCCGCACATCCTGCCGCCCACCGCCATCCGTCCTCTCGTGCTCGACCGCCAACGCTCTCTGCCGCACCGACCGCAGCCTGACCAGCAAACACTGCCCACTTCAATATCGCTGCCGATCTCGATAGCCGCTGTTCCCGAAGAACGGAAGGACCAAAAGAAAGAGCCCAGAGCACCTCCCATATCATTCCAAGTAACTCCTCCTGAGGGGAGCTGCCCTTTACTAGTGTTCATAAACCCTAAGAGCGGAGGGCGACAAGGGTCCAGGGTTCTAAGGAAACTCCAATATATCTTGAACCCGAGACAGGTTCATGATATCACCAAAGGAGGACCTACACAAGGTCTGCAGATGTTCAAAGACGTGAAGAACTATCGTGTCATATGCTGCGGTGGAGACGGTACAGTTGGCTGGGTTCTGGAGACCATGGATAAGGTGCAAATGGAAACTCAGCCGGCGGTCGGGGTGATACCTCTAGGAACCGGAAATGATCTGGCGCGGTGTCTGCGGTGGGGAGGAGGTTATGAAGGTGAATCGATTCACAAAATACTGGACAAGATCGCCAGAGCGAGCACTGTGATGATGGACAGGTGGCAGATTATGGTGGAAAACTTGGCCATGGAGTGTGATCAGCCTCAAATGCCAGACTCGGCGCCTCATCCCACAACAGTGCCGTACAACATCATCAATAATTATTTCTCCATCGGCGTTGACGCCGCCATCTGCGTGAAGTTCCACACGGAGAGAGAAAGGAATCCGGACAAGTTCAGTTCTAGAATGAAGAACAAGCTGTGGTACTTTGAGTTCGCGACGTCGGAACAGTTCGCAGCCAGCTGCAAGAACTTACACGAGAACATCGATATTGTGTGCGACGGGTCGTCTCTTGAGCTGAAGAAGGGTCCGTCGCTGCAGGGCGTGGCGCTCCTCAACATCCCGTACGCGCACGGCGGCTCCAACCTGTGGGGCACCAGCGCGGCGCAGCGGCGGGGGCGCTTCCCGCAAGCGCAGCAAGACATCGGCGACAGAATGATCGAAGTGATCGGACTTGAGAATTGTCTACACATGGGTCAGGTGCGGACAGGCCTTCGAGCCTCCGGGCGAAGGTTGGCCCAGTGCAGCTCAATAACGATCACCACGAAGAGAATGTTCCCGATGCAGATTGACGGCGAGCCCTGGATGCAGCCTCCGTGCACGATCACAATCACTCATAAAAATCAAGTTCCGATGCTCATGGGACCAGAGCCAGAGAAAGGAAGAGGATTCTTCAAAATATTCACTCACTGTTAA
Protein
MPNPVGGGFQWDKLSPAEFQQLQELASYSTRKLQDVLNEFCEVKKQSPDGDIDYEGFRWFLDTFLEVSTPDELSRHLFLSFVRRDRRPALREMAAASSTTACAAVTAHADHQVKPTLASKIHGLAERLQQLGKSSADSVDGSDKGSRSRTGSVHPMFTVTTHPSYSSHELCLDRRNETSPSHSQMSRNSSRKSSNSMRVNQSTKIEDFRHLMRRSSTVEVHTARVSLKDIVCYLSLLEAGRPEDKLEFMFRLYDTDGNGVLDTNEMDCIVNQMMAVAEYLGWETSELRPILQDMMVEIDYDADGTVSLDEWKRGGMTTIPLLVLLGLDTNVKEDGEHAWRLKHFSRPAYCNLCLNMLVGLGKKGLCCIFCKFTVHERCVQRAPASCIATYSKSKRASATLAHHWIEGNCHGKCARCRKKIKGYNGITGLHCRWCHITLHNRCVGAAGGACTLGRHAPHILPPTAIRPLVLDRQRSLPHRPQPDQQTLPTSISLPISIAAVPEERKDQKKEPRAPPISFQVTPPEGSCPLLVFINPKSGGRQGSRVLRKLQYILNPRQVHDITKGGPTQGLQMFKDVKNYRVICCGGDGTVGWVLETMDKVQMETQPAVGVIPLGTGNDLARCLRWGGGYEGESIHKILDKIARASTVMMDRWQIMVENLAMECDQPQMPDSAPHPTTVPYNIINNYFSIGVDAAICVKFHTERERNPDKFSSRMKNKLWYFEFATSEQFAASCKNLHENIDIVCDGSSLELKKGPSLQGVALLNIPYAHGGSNLWGTSAAQRRGRFPQAQQDIGDRMIEVIGLENCLHMGQVRTGLRASGRRLAQCSSITITTKRMFPMQIDGEPWMQPPCTITITHKNQVPMLMGPEPEKGRGFFKIFTHC
Summary
Description
Exhibits high phosphorylation activity for long-chain diacylglycerols.
Catalytic Activity
a 1,2-diacyl-sn-glycerol + ATP = a 1,2-diacyl-sn-glycero-3-phosphate + ADP + H(+)
Similarity
Belongs to the eukaryotic diacylglycerol kinase family.
Keywords
Alternative splicing
ATP-binding
Calcium
Complete proteome
Cytoplasm
Kinase
Metal-binding
Nucleotide-binding
Phosphoprotein
Polymorphism
Reference proteome
Repeat
Transferase
Zinc
Zinc-finger
Feature
chain Diacylglycerol kinase beta
splice variant In isoform 2.
sequence variant In dbSNP:rs34616903.
splice variant In isoform 2.
sequence variant In dbSNP:rs34616903.
Uniprot
A0A194PZ99
A0A194R029
A0A0K2S343
H9JUX3
A0A2A4J370
A0A087ZMS3
+ More
A0A3L8DCW2 A0A158NTM4 F4WSF1 A0A2A4J7Q7 A0A154PGA3 A0A1W4XJJ5 A0A1W4XKP6 A0A310SBY0 A0A026WJE9 A0A195ERV3 A0A1W4XJK0 A0A0L7RC29 A0A2A3E5B7 A0A1Y1K6F7 A0A151X3Q3 E2AXF7 A0A067RCJ5 A0A195BQ84 T1J4I9 A0A1W4XKG8 E2BKU8 T1ID18 E0VZ55 A0A1Y1KA40 A0A0P5XEF8 A0A1W7RAP3 A0A0P6H8X4 U4UEK8 E9H738 A0A1D2MP71 A0A210Q1M3 A0A1S3IRW2 N6UGH4 A0A2T7PUZ1 A0A1S3IPX2 A0A2R5L6H7 A0A1S3IQC2 E9IYH8 A0A1D1URY8 A0A1W0WBN0 A0A1W0WBI2 A0A0P5KC03 R7TZB0 A0A1S3IPW8 A0A1S3IQS8 A0A0P6C0W9 A0A0N8BDM6 A0A091HYU1 I3JHD4 A0A2Y9DMN4 A0A3P8NU32 A0A2Y9R8W3 F6ZFB7 A0A2K6C2Q0 A0A2K5KKT8 A0A2K5KKT3 A0A3P8NUA1 A0A2Y9DMK8 A0A337SXK4 A0A099Z8E5 A0A218V7R6 A0A3B4T403 A0A093IHD0 H0YWD4 B5MBY2 B7Z9A9 A0A3B4T4C0 U3K5V3 A0A091F352 A0A2Y9R8C8 A0A1S3FTV9 A0A287CTF2 G3RD41 A0A2U4B1M9 A0A2Y9T240 A0A2Y9H5U1 A0A2Y9H7J9 A0A2U3XIE8 A0A2U3VXH8 Q9Y6T7 Q6NS52-2 A0A2I2ZRH6 G1MD26 A0A2I3G9Q7 A0A2Y9DMN8 A0A2R9BEZ0 A0A2Y9H156 A0A2Y9JH57 A0A2Y9JMC4 H2RDM1 K1QAV3 B5MCD5
A0A3L8DCW2 A0A158NTM4 F4WSF1 A0A2A4J7Q7 A0A154PGA3 A0A1W4XJJ5 A0A1W4XKP6 A0A310SBY0 A0A026WJE9 A0A195ERV3 A0A1W4XJK0 A0A0L7RC29 A0A2A3E5B7 A0A1Y1K6F7 A0A151X3Q3 E2AXF7 A0A067RCJ5 A0A195BQ84 T1J4I9 A0A1W4XKG8 E2BKU8 T1ID18 E0VZ55 A0A1Y1KA40 A0A0P5XEF8 A0A1W7RAP3 A0A0P6H8X4 U4UEK8 E9H738 A0A1D2MP71 A0A210Q1M3 A0A1S3IRW2 N6UGH4 A0A2T7PUZ1 A0A1S3IPX2 A0A2R5L6H7 A0A1S3IQC2 E9IYH8 A0A1D1URY8 A0A1W0WBN0 A0A1W0WBI2 A0A0P5KC03 R7TZB0 A0A1S3IPW8 A0A1S3IQS8 A0A0P6C0W9 A0A0N8BDM6 A0A091HYU1 I3JHD4 A0A2Y9DMN4 A0A3P8NU32 A0A2Y9R8W3 F6ZFB7 A0A2K6C2Q0 A0A2K5KKT8 A0A2K5KKT3 A0A3P8NUA1 A0A2Y9DMK8 A0A337SXK4 A0A099Z8E5 A0A218V7R6 A0A3B4T403 A0A093IHD0 H0YWD4 B5MBY2 B7Z9A9 A0A3B4T4C0 U3K5V3 A0A091F352 A0A2Y9R8C8 A0A1S3FTV9 A0A287CTF2 G3RD41 A0A2U4B1M9 A0A2Y9T240 A0A2Y9H5U1 A0A2Y9H7J9 A0A2U3XIE8 A0A2U3VXH8 Q9Y6T7 Q6NS52-2 A0A2I2ZRH6 G1MD26 A0A2I3G9Q7 A0A2Y9DMN8 A0A2R9BEZ0 A0A2Y9H156 A0A2Y9JH57 A0A2Y9JMC4 H2RDM1 K1QAV3 B5MCD5
EC Number
2.7.1.107
Pubmed
EMBL
KQ459593
KPI96480.1
KQ460890
KPJ10889.1
AB999997
BAS21453.1
+ More
BABH01040760 BABH01040761 NWSH01003755 PCG65852.1 QOIP01000010 RLU17973.1 ADTU01026033 ADTU01026034 ADTU01026035 ADTU01026036 ADTU01026037 ADTU01026038 ADTU01026039 ADTU01026040 GL888321 EGI62868.1 NWSH01002674 PCG67698.1 KQ434899 KZC10881.1 KQ764145 OAD54595.1 KK107171 EZA56115.1 KQ981993 KYN30985.1 KQ414617 KOC68378.1 KZ288363 PBC26927.1 GEZM01091198 JAV57049.1 KQ982562 KYQ54848.1 GL443549 EFN61881.1 KK852727 KDR17623.1 KQ976424 KYM88670.1 JH431845 GL448826 EFN83697.1 ACPB03024605 ACPB03024606 DS235849 EEB18661.1 GEZM01091197 JAV57050.1 GDIP01074283 JAM29432.1 GFAH01000185 JAV48204.1 GDIQ01023013 JAN71724.1 KB632052 ERL88335.1 GL732599 EFX72450.1 LJIJ01000766 ODM94692.1 NEDP02005240 OWF42627.1 APGK01036200 APGK01036201 APGK01036202 APGK01036203 APGK01036204 APGK01036205 KB740937 ENN77757.1 PZQS01000002 PVD37197.1 GGLE01000911 MBY05037.1 GL767018 EFZ14349.1 BDGG01000001 GAU88928.1 MTYJ01000141 OQV12577.1 OQV12576.1 GDIQ01186446 JAK65279.1 AMQN01010248 KB307403 ELT98962.1 GDIP01007572 JAM96143.1 GDIQ01188051 JAK63674.1 KL218015 KFP01142.1 AERX01023956 AERX01023957 AERX01023958 AANG04003224 KL891452 KGL78779.1 MUZQ01000031 OWK62054.1 KL215714 KFV66151.1 ABQF01013448 ABQF01013449 ABQF01013450 ABQF01013451 ABQF01013452 ABQF01013453 AC005039 AC005248 AC006045 AC006150 AC011229 AC073258 AC105459 AK304719 BAH14245.1 AGTO01000092 AGTO01010212 KK718434 KFO56295.1 AGTP01054159 AGTP01054160 AGTP01054161 AGTP01054162 AGTP01054163 AGTP01054164 AGTP01054165 AGTP01054166 AGTP01054167 AGTP01054168 CABD030049949 CABD030049950 CABD030049951 CABD030049952 CABD030049953 CABD030049954 CABD030049955 CABD030049956 CABD030049957 CABD030049958 AX032742 CH236948 CH471073 BC105005 AB018261 AK139408 BC070461 AK122355 ACTA01115218 ACTA01123218 ACTA01131218 ACTA01139218 ACTA01147217 ACTA01155216 ACTA01163215 ACTA01171213 ACTA01179211 ACTA01187209 ADFV01001320 ADFV01001321 ADFV01001322 ADFV01001323 ADFV01001324 ADFV01001325 ADFV01001326 ADFV01001327 ADFV01001328 ADFV01001329 AJFE02088040 AJFE02088041 AJFE02088042 AJFE02088043 AJFE02088044 AJFE02088045 AJFE02088046 AJFE02088047 AJFE02088048 AJFE02088049 AACZ04068638 AACZ04068642 AC147975 AC188408 AC188412 AC190238 AC192126 NBAG03000259 PNI57915.1 JH818313 EKC31033.1
BABH01040760 BABH01040761 NWSH01003755 PCG65852.1 QOIP01000010 RLU17973.1 ADTU01026033 ADTU01026034 ADTU01026035 ADTU01026036 ADTU01026037 ADTU01026038 ADTU01026039 ADTU01026040 GL888321 EGI62868.1 NWSH01002674 PCG67698.1 KQ434899 KZC10881.1 KQ764145 OAD54595.1 KK107171 EZA56115.1 KQ981993 KYN30985.1 KQ414617 KOC68378.1 KZ288363 PBC26927.1 GEZM01091198 JAV57049.1 KQ982562 KYQ54848.1 GL443549 EFN61881.1 KK852727 KDR17623.1 KQ976424 KYM88670.1 JH431845 GL448826 EFN83697.1 ACPB03024605 ACPB03024606 DS235849 EEB18661.1 GEZM01091197 JAV57050.1 GDIP01074283 JAM29432.1 GFAH01000185 JAV48204.1 GDIQ01023013 JAN71724.1 KB632052 ERL88335.1 GL732599 EFX72450.1 LJIJ01000766 ODM94692.1 NEDP02005240 OWF42627.1 APGK01036200 APGK01036201 APGK01036202 APGK01036203 APGK01036204 APGK01036205 KB740937 ENN77757.1 PZQS01000002 PVD37197.1 GGLE01000911 MBY05037.1 GL767018 EFZ14349.1 BDGG01000001 GAU88928.1 MTYJ01000141 OQV12577.1 OQV12576.1 GDIQ01186446 JAK65279.1 AMQN01010248 KB307403 ELT98962.1 GDIP01007572 JAM96143.1 GDIQ01188051 JAK63674.1 KL218015 KFP01142.1 AERX01023956 AERX01023957 AERX01023958 AANG04003224 KL891452 KGL78779.1 MUZQ01000031 OWK62054.1 KL215714 KFV66151.1 ABQF01013448 ABQF01013449 ABQF01013450 ABQF01013451 ABQF01013452 ABQF01013453 AC005039 AC005248 AC006045 AC006150 AC011229 AC073258 AC105459 AK304719 BAH14245.1 AGTO01000092 AGTO01010212 KK718434 KFO56295.1 AGTP01054159 AGTP01054160 AGTP01054161 AGTP01054162 AGTP01054163 AGTP01054164 AGTP01054165 AGTP01054166 AGTP01054167 AGTP01054168 CABD030049949 CABD030049950 CABD030049951 CABD030049952 CABD030049953 CABD030049954 CABD030049955 CABD030049956 CABD030049957 CABD030049958 AX032742 CH236948 CH471073 BC105005 AB018261 AK139408 BC070461 AK122355 ACTA01115218 ACTA01123218 ACTA01131218 ACTA01139218 ACTA01147217 ACTA01155216 ACTA01163215 ACTA01171213 ACTA01179211 ACTA01187209 ADFV01001320 ADFV01001321 ADFV01001322 ADFV01001323 ADFV01001324 ADFV01001325 ADFV01001326 ADFV01001327 ADFV01001328 ADFV01001329 AJFE02088040 AJFE02088041 AJFE02088042 AJFE02088043 AJFE02088044 AJFE02088045 AJFE02088046 AJFE02088047 AJFE02088048 AJFE02088049 AACZ04068638 AACZ04068642 AC147975 AC188408 AC188412 AC190238 AC192126 NBAG03000259 PNI57915.1 JH818313 EKC31033.1
Proteomes
UP000053268
UP000053240
UP000005204
UP000218220
UP000005203
UP000279307
+ More
UP000005205 UP000007755 UP000076502 UP000192223 UP000053097 UP000078541 UP000053825 UP000242457 UP000075809 UP000000311 UP000027135 UP000078540 UP000008237 UP000015103 UP000009046 UP000030742 UP000000305 UP000094527 UP000242188 UP000085678 UP000019118 UP000245119 UP000186922 UP000014760 UP000054308 UP000005207 UP000248480 UP000265100 UP000008225 UP000233120 UP000233060 UP000011712 UP000053641 UP000197619 UP000261420 UP000053875 UP000007754 UP000005640 UP000016665 UP000052976 UP000081671 UP000005215 UP000001519 UP000245320 UP000248484 UP000248481 UP000245341 UP000245340 UP000000589 UP000008912 UP000001073 UP000240080 UP000248482 UP000002277 UP000005408
UP000005205 UP000007755 UP000076502 UP000192223 UP000053097 UP000078541 UP000053825 UP000242457 UP000075809 UP000000311 UP000027135 UP000078540 UP000008237 UP000015103 UP000009046 UP000030742 UP000000305 UP000094527 UP000242188 UP000085678 UP000019118 UP000245119 UP000186922 UP000014760 UP000054308 UP000005207 UP000248480 UP000265100 UP000008225 UP000233120 UP000233060 UP000011712 UP000053641 UP000197619 UP000261420 UP000053875 UP000007754 UP000005640 UP000016665 UP000052976 UP000081671 UP000005215 UP000001519 UP000245320 UP000248484 UP000248481 UP000245341 UP000245340 UP000000589 UP000008912 UP000001073 UP000240080 UP000248482 UP000002277 UP000005408
Pfam
Interpro
IPR001206
Diacylglycerol_kinase_cat_dom
+ More
IPR018247 EF_Hand_1_Ca_BS
IPR029477 DAG_kinase_typeI_N
IPR000756 Diacylglycerol_kin_accessory
IPR038199 DGK_typeI_N_sf
IPR002219 PE/DAG-bd
IPR037607 DGK
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR017438 ATP-NAD_kinase_N
IPR016064 NAD/diacylglycerol_kinase_sf
IPR018247 EF_Hand_1_Ca_BS
IPR029477 DAG_kinase_typeI_N
IPR000756 Diacylglycerol_kin_accessory
IPR038199 DGK_typeI_N_sf
IPR002219 PE/DAG-bd
IPR037607 DGK
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR017438 ATP-NAD_kinase_N
IPR016064 NAD/diacylglycerol_kinase_sf
Gene 3D
ProteinModelPortal
A0A194PZ99
A0A194R029
A0A0K2S343
H9JUX3
A0A2A4J370
A0A087ZMS3
+ More
A0A3L8DCW2 A0A158NTM4 F4WSF1 A0A2A4J7Q7 A0A154PGA3 A0A1W4XJJ5 A0A1W4XKP6 A0A310SBY0 A0A026WJE9 A0A195ERV3 A0A1W4XJK0 A0A0L7RC29 A0A2A3E5B7 A0A1Y1K6F7 A0A151X3Q3 E2AXF7 A0A067RCJ5 A0A195BQ84 T1J4I9 A0A1W4XKG8 E2BKU8 T1ID18 E0VZ55 A0A1Y1KA40 A0A0P5XEF8 A0A1W7RAP3 A0A0P6H8X4 U4UEK8 E9H738 A0A1D2MP71 A0A210Q1M3 A0A1S3IRW2 N6UGH4 A0A2T7PUZ1 A0A1S3IPX2 A0A2R5L6H7 A0A1S3IQC2 E9IYH8 A0A1D1URY8 A0A1W0WBN0 A0A1W0WBI2 A0A0P5KC03 R7TZB0 A0A1S3IPW8 A0A1S3IQS8 A0A0P6C0W9 A0A0N8BDM6 A0A091HYU1 I3JHD4 A0A2Y9DMN4 A0A3P8NU32 A0A2Y9R8W3 F6ZFB7 A0A2K6C2Q0 A0A2K5KKT8 A0A2K5KKT3 A0A3P8NUA1 A0A2Y9DMK8 A0A337SXK4 A0A099Z8E5 A0A218V7R6 A0A3B4T403 A0A093IHD0 H0YWD4 B5MBY2 B7Z9A9 A0A3B4T4C0 U3K5V3 A0A091F352 A0A2Y9R8C8 A0A1S3FTV9 A0A287CTF2 G3RD41 A0A2U4B1M9 A0A2Y9T240 A0A2Y9H5U1 A0A2Y9H7J9 A0A2U3XIE8 A0A2U3VXH8 Q9Y6T7 Q6NS52-2 A0A2I2ZRH6 G1MD26 A0A2I3G9Q7 A0A2Y9DMN8 A0A2R9BEZ0 A0A2Y9H156 A0A2Y9JH57 A0A2Y9JMC4 H2RDM1 K1QAV3 B5MCD5
A0A3L8DCW2 A0A158NTM4 F4WSF1 A0A2A4J7Q7 A0A154PGA3 A0A1W4XJJ5 A0A1W4XKP6 A0A310SBY0 A0A026WJE9 A0A195ERV3 A0A1W4XJK0 A0A0L7RC29 A0A2A3E5B7 A0A1Y1K6F7 A0A151X3Q3 E2AXF7 A0A067RCJ5 A0A195BQ84 T1J4I9 A0A1W4XKG8 E2BKU8 T1ID18 E0VZ55 A0A1Y1KA40 A0A0P5XEF8 A0A1W7RAP3 A0A0P6H8X4 U4UEK8 E9H738 A0A1D2MP71 A0A210Q1M3 A0A1S3IRW2 N6UGH4 A0A2T7PUZ1 A0A1S3IPX2 A0A2R5L6H7 A0A1S3IQC2 E9IYH8 A0A1D1URY8 A0A1W0WBN0 A0A1W0WBI2 A0A0P5KC03 R7TZB0 A0A1S3IPW8 A0A1S3IQS8 A0A0P6C0W9 A0A0N8BDM6 A0A091HYU1 I3JHD4 A0A2Y9DMN4 A0A3P8NU32 A0A2Y9R8W3 F6ZFB7 A0A2K6C2Q0 A0A2K5KKT8 A0A2K5KKT3 A0A3P8NUA1 A0A2Y9DMK8 A0A337SXK4 A0A099Z8E5 A0A218V7R6 A0A3B4T403 A0A093IHD0 H0YWD4 B5MBY2 B7Z9A9 A0A3B4T4C0 U3K5V3 A0A091F352 A0A2Y9R8C8 A0A1S3FTV9 A0A287CTF2 G3RD41 A0A2U4B1M9 A0A2Y9T240 A0A2Y9H5U1 A0A2Y9H7J9 A0A2U3XIE8 A0A2U3VXH8 Q9Y6T7 Q6NS52-2 A0A2I2ZRH6 G1MD26 A0A2I3G9Q7 A0A2Y9DMN8 A0A2R9BEZ0 A0A2Y9H156 A0A2Y9JH57 A0A2Y9JMC4 H2RDM1 K1QAV3 B5MCD5
PDB
6IIE
E-value=1.54725e-33,
Score=361
Ontologies
PATHWAY
GO
PANTHER
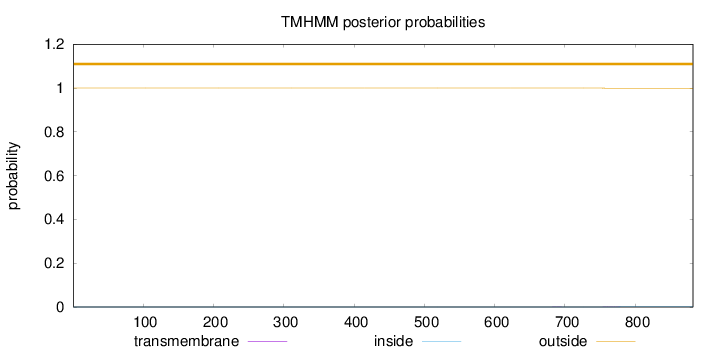
Topology
Subcellular location
Cytoplasm
Length:
882
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02176
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00005
outside
1 - 882
Population Genetic Test Statistics
Pi
210.767856
Theta
151.362032
Tajima's D
1.386417
CLR
0.04074
CSRT
0.757662116894155
Interpretation
Uncertain
