Gene
KWMTBOMO00731
Pre Gene Modal
BGIBMGA003253
Annotation
PREDICTED:_ITG-containing_peptide_isoform_X1_[Bombyx_mori]
Full name
ITG-like peptide
+ More
Prohormone-3
Prohormone-3
Location in the cell
Extracellular Reliability : 4.067
Sequence
CDS
ATGTCTCATTTCGCCATGACTGCCACCGCGATCTTAGTGCTGGGCTGTCTCTCCGGCGCCCACGCCTGGGGTGGCCTCTTCAACCGTTTCAGCTCCGATATGCTGGCCAATCTTGGCTACGGAAGGTCCCCGTATCGCCACTATCCTTACGGGCAGGTGGAACCTGACGAAGCGTACGAGGCCCTGGAAAACAACCGCATCTCGAACGTGATTGATGAACCAGCACACTGCTATAGCTCCCCGTGTGTGACCAACGGCGATTGCTGCGGCGGACTTCTGTGTCTGGAGACCGATGACGGAGGACGGTGCCTATCAGCGTTTGCTGGAAGGAAGCTTGGCGAGATCTGCAACCGAGAGAACCAATGCGATGCCGGACTGATCTGCGAGGAAGCTGCTCCTGGTGAGATGCACATCTGCCGCCCGCCTTCCACCGGCCGCAAGCAATACAACGAGGACTGCACCACGTCCAGCGAATGCGAGATCACCAGGGGACTCTGCTGCATCATGCAACGCCGACACCGCCAGAAGTCTAGGAAGAGCTGTGGATACTTCAAGGAGCCCCTGGTCTGCATCGGCCCGGTGGCCATCGACCAGATCCGCGAGTACGTGGAGCACACGACCGGCGAAAAGCGCATCGGAGCTTACCGCCTCCATTAG
Protein
MSHFAMTATAILVLGCLSGAHAWGGLFNRFSSDMLANLGYGRSPYRHYPYGQVEPDEAYEALENNRISNVIDEPAHCYSSPCVTNGDCCGGLLCLETDDGGRCLSAFAGRKLGEICNRENQCDAGLICEEAAPGEMHICRPPSTGRKQYNEDCTTSSECEITRGLCCIMQRRHRQKSRKSCGYFKEPLVCIGPVAIDQIREYVEHTTGEKRIGAYRLH
Summary
Similarity
Belongs to the peptidase S1 family.
Keywords
Cleavage on pair of basic residues
Direct protein sequencing
Neuropeptide
Secreted
Signal
Complete proteome
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
peptide ITG-like peptide
chain Prohormone-3
chain Prohormone-3
Uniprot
B3IUC9
A0A2A4K029
M4QNS2
A0A0S1U091
C0HKU1
A0A212EXW3
+ More
A0A194PUT9 A0A194R501 A0A336LQ05 A0A1Y1MF30 T1DF17 B0WE25 A0A023EJR4 A0A067RK19 A0A1Q3FRA5 A0A182NZG5 A0A1S4H0L2 A0A1Q3FRC4 A0A182QVC7 Q16TD8 Q7PWE6 A0A182RHC4 U5EU83 A0A232ELX3 A0A348G697 A0A182F5Y7 K7INL7 A0A182WGD8 A0A182LMH7 A0A182IZK8 A0A182TBW0 A0A1W6EVR2 A0A1D2MKY4 A0A0N1IT68 A0A182UW25 T1D4W8 A0A139WEY0 E9IVI7 A0A226EG08 F4WIU0 A0A151WLF9 A0A0L7R6R4 A0A0C9QE28 A0A195B4F3 A0A158P492 A0A3L8E1U9 A0A151IXP6 A0A195CTX7 A0A026VUH6 A0A1J1IEF9 A0A182Y0J1 A0A2A3E9M2 V9IJ72 A0A1B6K7P4 A0A1B6ENU8 A0A088AGP0 A0A1W4WX08 A0A154P565 A0A2R7WFR3 P85828 E2C4I5 N6SSK4 A0A0A9YIV8 A0A2U9PFR8 A0A162RH31 A0A0P6ABD5 A0A0P6I1W0 E2ADG2 A0A2P8Y705 A0A1B6KBK9 A0A1B6CVC5 A0A1B6E6L7 A0A182M0I7 A0A1S3D513 A0A0K2T3B5 E9H740 A0A182TG45 A0A0P5AEL0 A0A0J7KGC9 A0A182NBL3 A0A0P5BEQ1 T1J3R1 A0A0A1XPZ0
A0A194PUT9 A0A194R501 A0A336LQ05 A0A1Y1MF30 T1DF17 B0WE25 A0A023EJR4 A0A067RK19 A0A1Q3FRA5 A0A182NZG5 A0A1S4H0L2 A0A1Q3FRC4 A0A182QVC7 Q16TD8 Q7PWE6 A0A182RHC4 U5EU83 A0A232ELX3 A0A348G697 A0A182F5Y7 K7INL7 A0A182WGD8 A0A182LMH7 A0A182IZK8 A0A182TBW0 A0A1W6EVR2 A0A1D2MKY4 A0A0N1IT68 A0A182UW25 T1D4W8 A0A139WEY0 E9IVI7 A0A226EG08 F4WIU0 A0A151WLF9 A0A0L7R6R4 A0A0C9QE28 A0A195B4F3 A0A158P492 A0A3L8E1U9 A0A151IXP6 A0A195CTX7 A0A026VUH6 A0A1J1IEF9 A0A182Y0J1 A0A2A3E9M2 V9IJ72 A0A1B6K7P4 A0A1B6ENU8 A0A088AGP0 A0A1W4WX08 A0A154P565 A0A2R7WFR3 P85828 E2C4I5 N6SSK4 A0A0A9YIV8 A0A2U9PFR8 A0A162RH31 A0A0P6ABD5 A0A0P6I1W0 E2ADG2 A0A2P8Y705 A0A1B6KBK9 A0A1B6CVC5 A0A1B6E6L7 A0A182M0I7 A0A1S3D513 A0A0K2T3B5 E9H740 A0A182TG45 A0A0P5AEL0 A0A0J7KGC9 A0A182NBL3 A0A0P5BEQ1 T1J3R1 A0A0A1XPZ0
Pubmed
28756777
29466015
22118469
26354079
28004739
24330624
+ More
24945155 24845553 12364791 17510324 14747013 17210077 28648823 20075255 20966253 27289101 18362917 19820115 21282665 21719571 21347285 30249741 24508170 25244985 17068263 20798317 23537049 25401762 26823975 25641051 29403074 21292972 25830018
24945155 24845553 12364791 17510324 14747013 17210077 28648823 20075255 20966253 27289101 18362917 19820115 21282665 21719571 21347285 30249741 24508170 25244985 17068263 20798317 23537049 25401762 26823975 25641051 29403074 21292972 25830018
EMBL
AB298925
BAG50364.1
NWSH01000343
PCG77254.1
KC340916
KZ150065
+ More
AGH25548.1 PZC74170.1 KT005954 RSAL01000343 ALM30305.1 RVE42372.1 AGBW02011664 OWR46333.1 KQ459593 KPI96514.1 KQ460890 KPJ10941.1 UFQT01000101 SSX20000.1 GEZM01036151 JAV82935.1 GALA01000837 JAA94015.1 DS231904 EDS45201.1 GAPW01004301 JAC09297.1 KK852427 KDR24122.1 GFDL01005039 JAV30006.1 AAAB01008984 GFDL01005019 JAV30026.1 AXCN02000676 CH477651 CH477387 EAT37778.1 EAT42061.1 EAA15065.4 GANO01004000 JAB55871.1 NNAY01003486 OXU19346.1 FX986013 BBF97970.1 KY563409 ARK19818.1 LJIJ01000934 ODM93680.1 KQ435896 KOX69321.1 GALA01000798 JAA94054.1 KQ971354 KYB26538.1 GL766258 EFZ15401.1 LNIX01000004 OXA56482.1 GL888177 EGI65886.1 KQ982959 KYQ48698.1 KQ414646 KOC66519.1 GBYB01001568 GBYB01001569 JAG71335.1 JAG71336.1 KQ976618 KYM79160.1 ADTU01008748 QOIP01000001 RLU26545.1 RLU26606.1 KQ980796 KYN13007.1 KQ977279 KYN04143.1 KK107928 EZA47131.1 CVRI01000047 CRK97940.1 KZ288326 PBC27982.1 JR050062 AEY61168.1 GECU01010095 GECU01004808 GECU01000228 JAS97611.1 JAT02899.1 JAT07479.1 GECZ01030163 GECZ01029027 GECZ01014598 GECZ01009453 GECZ01009079 JAS39606.1 JAS40742.1 JAS55171.1 JAS60316.1 JAS60690.1 KQ434822 KZC07076.1 KK854766 PTY18507.1 PTY18508.1 AADG06002762 GL452471 EFN77169.1 APGK01058247 APGK01058248 KB741288 KB631652 ENN70639.1 ERL85038.1 GBHO01012571 GBRD01015959 GDHC01000662 JAG31033.1 JAG49867.1 JAQ17967.1 MG550172 AWT50606.1 LRGB01000190 KZS20504.1 GDIP01032909 JAM70806.1 GDIQ01010721 JAN84016.1 GL438750 EFN68524.1 PYGN01000852 PSN40033.1 GEBQ01031146 JAT08831.1 GEDC01020013 GEDC01019744 JAS17285.1 JAS17554.1 GEDC01016003 GEDC01003730 JAS21295.1 JAS33568.1 AXCM01003792 HACA01002550 CDW19911.1 GL732599 EFX72509.1 GDIP01200847 JAJ22555.1 LBMM01007727 KMQ89478.1 GDIP01200846 JAJ22556.1 JH431830 GBXI01013696 GBXI01001196 JAD00596.1 JAD13096.1
AGH25548.1 PZC74170.1 KT005954 RSAL01000343 ALM30305.1 RVE42372.1 AGBW02011664 OWR46333.1 KQ459593 KPI96514.1 KQ460890 KPJ10941.1 UFQT01000101 SSX20000.1 GEZM01036151 JAV82935.1 GALA01000837 JAA94015.1 DS231904 EDS45201.1 GAPW01004301 JAC09297.1 KK852427 KDR24122.1 GFDL01005039 JAV30006.1 AAAB01008984 GFDL01005019 JAV30026.1 AXCN02000676 CH477651 CH477387 EAT37778.1 EAT42061.1 EAA15065.4 GANO01004000 JAB55871.1 NNAY01003486 OXU19346.1 FX986013 BBF97970.1 KY563409 ARK19818.1 LJIJ01000934 ODM93680.1 KQ435896 KOX69321.1 GALA01000798 JAA94054.1 KQ971354 KYB26538.1 GL766258 EFZ15401.1 LNIX01000004 OXA56482.1 GL888177 EGI65886.1 KQ982959 KYQ48698.1 KQ414646 KOC66519.1 GBYB01001568 GBYB01001569 JAG71335.1 JAG71336.1 KQ976618 KYM79160.1 ADTU01008748 QOIP01000001 RLU26545.1 RLU26606.1 KQ980796 KYN13007.1 KQ977279 KYN04143.1 KK107928 EZA47131.1 CVRI01000047 CRK97940.1 KZ288326 PBC27982.1 JR050062 AEY61168.1 GECU01010095 GECU01004808 GECU01000228 JAS97611.1 JAT02899.1 JAT07479.1 GECZ01030163 GECZ01029027 GECZ01014598 GECZ01009453 GECZ01009079 JAS39606.1 JAS40742.1 JAS55171.1 JAS60316.1 JAS60690.1 KQ434822 KZC07076.1 KK854766 PTY18507.1 PTY18508.1 AADG06002762 GL452471 EFN77169.1 APGK01058247 APGK01058248 KB741288 KB631652 ENN70639.1 ERL85038.1 GBHO01012571 GBRD01015959 GDHC01000662 JAG31033.1 JAG49867.1 JAQ17967.1 MG550172 AWT50606.1 LRGB01000190 KZS20504.1 GDIP01032909 JAM70806.1 GDIQ01010721 JAN84016.1 GL438750 EFN68524.1 PYGN01000852 PSN40033.1 GEBQ01031146 JAT08831.1 GEDC01020013 GEDC01019744 JAS17285.1 JAS17554.1 GEDC01016003 GEDC01003730 JAS21295.1 JAS33568.1 AXCM01003792 HACA01002550 CDW19911.1 GL732599 EFX72509.1 GDIP01200847 JAJ22555.1 LBMM01007727 KMQ89478.1 GDIP01200846 JAJ22556.1 JH431830 GBXI01013696 GBXI01001196 JAD00596.1 JAD13096.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
UP000002320
+ More
UP000027135 UP000075885 UP000075886 UP000008820 UP000007062 UP000075900 UP000215335 UP000069272 UP000002358 UP000075920 UP000075882 UP000075880 UP000075901 UP000094527 UP000053105 UP000075903 UP000007266 UP000198287 UP000007755 UP000075809 UP000053825 UP000078540 UP000005205 UP000279307 UP000078492 UP000078542 UP000053097 UP000183832 UP000076408 UP000242457 UP000005203 UP000192223 UP000076502 UP000008237 UP000019118 UP000030742 UP000076858 UP000000311 UP000245037 UP000075883 UP000079169 UP000000305 UP000075902 UP000036403 UP000075884
UP000027135 UP000075885 UP000075886 UP000008820 UP000007062 UP000075900 UP000215335 UP000069272 UP000002358 UP000075920 UP000075882 UP000075880 UP000075901 UP000094527 UP000053105 UP000075903 UP000007266 UP000198287 UP000007755 UP000075809 UP000053825 UP000078540 UP000005205 UP000279307 UP000078492 UP000078542 UP000053097 UP000183832 UP000076408 UP000242457 UP000005203 UP000192223 UP000076502 UP000008237 UP000019118 UP000030742 UP000076858 UP000000311 UP000245037 UP000075883 UP000079169 UP000000305 UP000075902 UP000036403 UP000075884
PRIDE
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
B3IUC9
A0A2A4K029
M4QNS2
A0A0S1U091
C0HKU1
A0A212EXW3
+ More
A0A194PUT9 A0A194R501 A0A336LQ05 A0A1Y1MF30 T1DF17 B0WE25 A0A023EJR4 A0A067RK19 A0A1Q3FRA5 A0A182NZG5 A0A1S4H0L2 A0A1Q3FRC4 A0A182QVC7 Q16TD8 Q7PWE6 A0A182RHC4 U5EU83 A0A232ELX3 A0A348G697 A0A182F5Y7 K7INL7 A0A182WGD8 A0A182LMH7 A0A182IZK8 A0A182TBW0 A0A1W6EVR2 A0A1D2MKY4 A0A0N1IT68 A0A182UW25 T1D4W8 A0A139WEY0 E9IVI7 A0A226EG08 F4WIU0 A0A151WLF9 A0A0L7R6R4 A0A0C9QE28 A0A195B4F3 A0A158P492 A0A3L8E1U9 A0A151IXP6 A0A195CTX7 A0A026VUH6 A0A1J1IEF9 A0A182Y0J1 A0A2A3E9M2 V9IJ72 A0A1B6K7P4 A0A1B6ENU8 A0A088AGP0 A0A1W4WX08 A0A154P565 A0A2R7WFR3 P85828 E2C4I5 N6SSK4 A0A0A9YIV8 A0A2U9PFR8 A0A162RH31 A0A0P6ABD5 A0A0P6I1W0 E2ADG2 A0A2P8Y705 A0A1B6KBK9 A0A1B6CVC5 A0A1B6E6L7 A0A182M0I7 A0A1S3D513 A0A0K2T3B5 E9H740 A0A182TG45 A0A0P5AEL0 A0A0J7KGC9 A0A182NBL3 A0A0P5BEQ1 T1J3R1 A0A0A1XPZ0
A0A194PUT9 A0A194R501 A0A336LQ05 A0A1Y1MF30 T1DF17 B0WE25 A0A023EJR4 A0A067RK19 A0A1Q3FRA5 A0A182NZG5 A0A1S4H0L2 A0A1Q3FRC4 A0A182QVC7 Q16TD8 Q7PWE6 A0A182RHC4 U5EU83 A0A232ELX3 A0A348G697 A0A182F5Y7 K7INL7 A0A182WGD8 A0A182LMH7 A0A182IZK8 A0A182TBW0 A0A1W6EVR2 A0A1D2MKY4 A0A0N1IT68 A0A182UW25 T1D4W8 A0A139WEY0 E9IVI7 A0A226EG08 F4WIU0 A0A151WLF9 A0A0L7R6R4 A0A0C9QE28 A0A195B4F3 A0A158P492 A0A3L8E1U9 A0A151IXP6 A0A195CTX7 A0A026VUH6 A0A1J1IEF9 A0A182Y0J1 A0A2A3E9M2 V9IJ72 A0A1B6K7P4 A0A1B6ENU8 A0A088AGP0 A0A1W4WX08 A0A154P565 A0A2R7WFR3 P85828 E2C4I5 N6SSK4 A0A0A9YIV8 A0A2U9PFR8 A0A162RH31 A0A0P6ABD5 A0A0P6I1W0 E2ADG2 A0A2P8Y705 A0A1B6KBK9 A0A1B6CVC5 A0A1B6E6L7 A0A182M0I7 A0A1S3D513 A0A0K2T3B5 E9H740 A0A182TG45 A0A0P5AEL0 A0A0J7KGC9 A0A182NBL3 A0A0P5BEQ1 T1J3R1 A0A0A1XPZ0
Ontologies
Topology
Subcellular location
Secreted
Membrane
Membrane
SignalP
Position: 1 - 22,
Likelihood: 0.977393
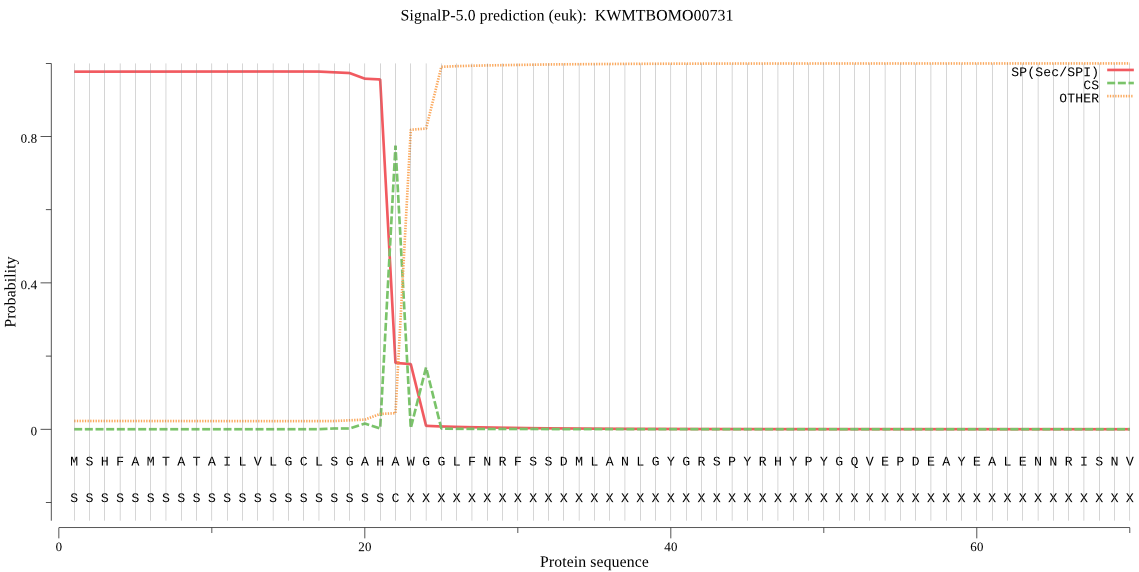
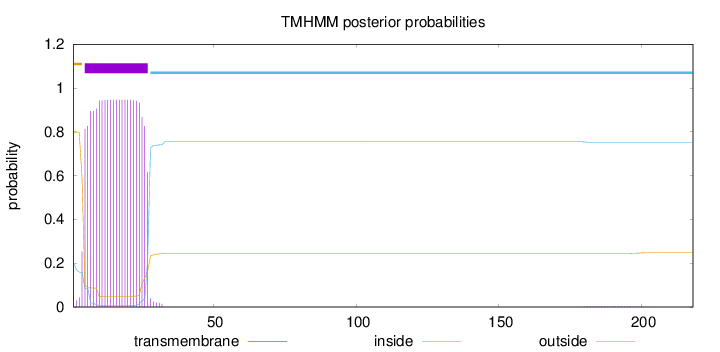
Length:
218
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.3054
Exp number, first 60 AAs:
21.24946
Total prob of N-in:
0.20189
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 27
inside
28 - 218
Population Genetic Test Statistics
Pi
253.89213
Theta
153.850162
Tajima's D
2.252294
CLR
0.032926
CSRT
0.91900404979751
Interpretation
Uncertain
