Gene
KWMTBOMO00728
Pre Gene Modal
BGIBMGA003257
Annotation
PREDICTED:_coiled-coil_domain-containing_protein_113-like_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 1.955
Sequence
CDS
ATGTCTCAAGGAGTCCACAGGAGTAGCATCAGCGCCACATCGGGCAGCCACTACGACGAACTCAGCGACACGGAGCTGATAAAGCTGGTCGGCGAATTGAAACAGAACCTCAAGGCGTTGCAACTCGAAAACGACGTACTCGAAAGGGTCCTGACGAGGCTGGACCCTTCCCTGCTGCAGGGCATCCAACAGGCCATCGAATACGCGAGCAAATTTCAGAGCTACTCCTCGATGAACGTGGGCAGTTTCGTCAAATCGCAAACTTCCAAATTCGGGTTCGAGTCCCTAGCTAGCCCATCCAGGATGCTCACCAGCCCTTCGAAAGTTTCGACCAGGCGAATAGACTCATCTGCCAGGGTCGGAGGGACGGTCATCTTCGGCACGGGACCACGAGTCAACGTGTTAGAGAGATCGGAATTAGTTTCCACCGAAATGGAGATCGTGATACGGAATCTCGAGAAAGTCCGGAGCAAGGCCGCTAAGCAACACGCTTTATTAAAAGCTCAATTAGAAGAGATAGGCCTAAGGGTCAATGATATCGTGAAATCATACGAGCTATTCGACAAGCAGGTTATCGTCGAAGGCTGGGACAGTATAGCCCAAAGGATCCCCGCTGAAATTTGGATACGATTCATGACTGAGTGGGTCAAAGCGACCGACAGCCAGATCGGGAAGCTGCGGCTCCGGACCAGCACCCTGAACACTCACTACAGCAAATTAAAAGGACAGATCCAAGTCAAAGAGGAGCTCAGCCAGAATCTAAGACCCGTCGATTTTGACAAACTGAAGATCGAAAACGGAGAATGCATTAGCGTTATCGAACAGAAGATTCAGCAGCTTGGCGAACTTAAGAAGATGACGGGCGACGCCAATTTGAATCTGGCCGTCCACAAGAAGGCTCTGATGCAGCAGAACACATACCTAGCCAACGTGTTGAGAACGATCAGAACGAAGCAGCAGCAAGCCGCGGTATTAGATAGAGAGAAGAATGTAATAGAAGACGAAGCGGATAAGCTGGAGGATAAGCTGGAGGGGGTGAGGAAATTGAGGTCAGCTTTCGAAGTGCCCGATGTCATGGACTACGTGAGGGTCAAAGCTGAGGTGGCAGACCTAAAGCACTCGATAAAGATGCTGGAAAACCGAAGCAGAGTACAGCAGTTCGCTTTGAAATCTTTGACCAGGCAATTGAAGATTTTGAATTGA
Protein
MSQGVHRSSISATSGSHYDELSDTELIKLVGELKQNLKALQLENDVLERVLTRLDPSLLQGIQQAIEYASKFQSYSSMNVGSFVKSQTSKFGFESLASPSRMLTSPSKVSTRRIDSSARVGGTVIFGTGPRVNVLERSELVSTEMEIVIRNLEKVRSKAAKQHALLKAQLEEIGLRVNDIVKSYELFDKQVIVEGWDSIAQRIPAEIWIRFMTEWVKATDSQIGKLRLRTSTLNTHYSKLKGQIQVKEELSQNLRPVDFDKLKIENGECISVIEQKIQQLGELKKMTGDANLNLAVHKKALMQQNTYLANVLRTIRTKQQQAAVLDREKNVIEDEADKLEDKLEGVRKLRSAFEVPDVMDYVRVKAEVADLKHSIKMLENRSRVQQFALKSLTRQLKILN
Summary
Uniprot
H9J170
A0A2A4JNF3
A0A194R507
A0A194PUU4
A0A2H1X1D1
A0A212FD22
+ More
A0A2A4JM89 A0A2H1WHX7 A0A0L7L9J8 A0A1W4XKN5 E2ASG8 A0A139WHB0 A0A026W8H0 E2BY35 E9IJW5 A0A195F7D1 A0A195D0B3 A0A3L8DI89 A0A195D9X7 A0A151WYE3 K7IUY2 A0A195B900 A0A232EUZ1 A0A2P8XL57 A0A0M8ZQS1 A0A0L0HK43 C3ZBE2 A0A1S3J674 A0A3M6UNB4 A7SJZ5 A0A2B4T1I5 V3YXT5 D2A2E0 A0A1B6DVC6 K1Q2F8 T2M5P4 A0A1B6LMP6 A0A210QH24 V9FKP8 W2XGN2 A0A081AMJ1 W2H7H4 A0A0W8D8M9 W2QGY5 W2ZPW0 W2LK35 A0A1S3MM96 A0A369SJG4 B3RVE1 D2A2D9 A0A2K5N0Z3 A0A060VXI0 A0A1W4Z333 A0A3B4C167 D0NZ21 A0A2K5Z423 A0A098M1B5 A0A2K6B738 A0A2K5K9W1 A0A2K6M7P5 A0A2K5UQZ9 A0A096NJ05 H9F3K1 A0A1S3MME4 A0A388KSS2 A0A0D9QXK7 A0A2U3XN24 W5M7M5 A0A2U3VLF9
A0A2A4JM89 A0A2H1WHX7 A0A0L7L9J8 A0A1W4XKN5 E2ASG8 A0A139WHB0 A0A026W8H0 E2BY35 E9IJW5 A0A195F7D1 A0A195D0B3 A0A3L8DI89 A0A195D9X7 A0A151WYE3 K7IUY2 A0A195B900 A0A232EUZ1 A0A2P8XL57 A0A0M8ZQS1 A0A0L0HK43 C3ZBE2 A0A1S3J674 A0A3M6UNB4 A7SJZ5 A0A2B4T1I5 V3YXT5 D2A2E0 A0A1B6DVC6 K1Q2F8 T2M5P4 A0A1B6LMP6 A0A210QH24 V9FKP8 W2XGN2 A0A081AMJ1 W2H7H4 A0A0W8D8M9 W2QGY5 W2ZPW0 W2LK35 A0A1S3MM96 A0A369SJG4 B3RVE1 D2A2D9 A0A2K5N0Z3 A0A060VXI0 A0A1W4Z333 A0A3B4C167 D0NZ21 A0A2K5Z423 A0A098M1B5 A0A2K6B738 A0A2K5K9W1 A0A2K6M7P5 A0A2K5UQZ9 A0A096NJ05 H9F3K1 A0A1S3MME4 A0A388KSS2 A0A0D9QXK7 A0A2U3XN24 W5M7M5 A0A2U3VLF9
Pubmed
EMBL
BABH01031368
NWSH01001044
PCG72940.1
KQ460890
KPJ10946.1
KQ459593
+ More
KPI96519.1 ODYU01012681 SOQ59121.1 AGBW02009127 OWR51598.1 PCG72939.1 ODYU01008776 SOQ52671.1 JTDY01002186 KOB71921.1 GL442298 EFN63634.1 KQ971343 KYB27306.1 KK107347 EZA52350.1 GL451412 EFN79408.1 GL763883 EFZ19157.1 KQ981744 KYN36348.1 KQ977012 KYN06358.1 QOIP01000007 RLU20036.1 KQ981135 KYN09209.1 KQ982650 KYQ52899.1 AAZX01001184 KQ976558 KYM80707.1 NNAY01002067 OXU22172.1 PYGN01001799 PSN32746.1 KQ435896 KOX69220.1 KQ257454 KND01403.1 GG666603 EEN50179.1 RCHS01001112 RMX55183.1 DS469682 EDO35964.1 LSMT01000004 PFX34485.1 KB203854 ESO82878.1 KQ971338 EFA02036.1 GEDC01007662 JAS29636.1 JH816822 EKC28118.1 HAAD01001222 CDG67454.1 GEBQ01014966 JAT25011.1 NEDP02003738 OWF48016.1 ANIZ01000930 ETI51353.1 ANIX01001130 ETP21129.1 ANJA01001038 ETO80102.1 KI685376 KI671936 ETK91243.1 ETL44643.1 LNFO01003985 LNFP01000461 KUF81795.1 KUF92536.1 KI669578 ETN11764.1 ANIY01001164 ETP49026.1 KI678669 KI691914 ETL97812.1 ETM50969.1 NOWV01000005 RDD46899.1 DS985244 EDV25978.1 EFA02037.1 FR904327 CDQ59511.1 DS028193 EEY68808.1 GBSI01000481 JAC96015.1 AQIA01043040 AQIA01043041 AQIA01043042 AHZZ02018805 JU325454 AFE69210.1 BFEA01000175 GBG72983.1 AQIB01137312 AHAT01010548
KPI96519.1 ODYU01012681 SOQ59121.1 AGBW02009127 OWR51598.1 PCG72939.1 ODYU01008776 SOQ52671.1 JTDY01002186 KOB71921.1 GL442298 EFN63634.1 KQ971343 KYB27306.1 KK107347 EZA52350.1 GL451412 EFN79408.1 GL763883 EFZ19157.1 KQ981744 KYN36348.1 KQ977012 KYN06358.1 QOIP01000007 RLU20036.1 KQ981135 KYN09209.1 KQ982650 KYQ52899.1 AAZX01001184 KQ976558 KYM80707.1 NNAY01002067 OXU22172.1 PYGN01001799 PSN32746.1 KQ435896 KOX69220.1 KQ257454 KND01403.1 GG666603 EEN50179.1 RCHS01001112 RMX55183.1 DS469682 EDO35964.1 LSMT01000004 PFX34485.1 KB203854 ESO82878.1 KQ971338 EFA02036.1 GEDC01007662 JAS29636.1 JH816822 EKC28118.1 HAAD01001222 CDG67454.1 GEBQ01014966 JAT25011.1 NEDP02003738 OWF48016.1 ANIZ01000930 ETI51353.1 ANIX01001130 ETP21129.1 ANJA01001038 ETO80102.1 KI685376 KI671936 ETK91243.1 ETL44643.1 LNFO01003985 LNFP01000461 KUF81795.1 KUF92536.1 KI669578 ETN11764.1 ANIY01001164 ETP49026.1 KI678669 KI691914 ETL97812.1 ETM50969.1 NOWV01000005 RDD46899.1 DS985244 EDV25978.1 EFA02037.1 FR904327 CDQ59511.1 DS028193 EEY68808.1 GBSI01000481 JAC96015.1 AQIA01043040 AQIA01043041 AQIA01043042 AHZZ02018805 JU325454 AFE69210.1 BFEA01000175 GBG72983.1 AQIB01137312 AHAT01010548
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000192223 UP000000311 UP000007266 UP000053097 UP000008237 UP000078541 UP000078542 UP000279307 UP000078492 UP000075809 UP000002358 UP000078540 UP000215335 UP000245037 UP000053105 UP000053201 UP000001554 UP000085678 UP000275408 UP000001593 UP000225706 UP000030746 UP000005408 UP000242188 UP000018721 UP000018958 UP000028582 UP000053236 UP000053864 UP000052943 UP000018817 UP000018948 UP000054423 UP000054532 UP000087266 UP000253843 UP000009022 UP000233060 UP000193380 UP000192224 UP000261440 UP000006643 UP000233140 UP000233120 UP000233080 UP000233180 UP000233100 UP000028761 UP000265515 UP000029965 UP000245341 UP000018468 UP000245340
UP000192223 UP000000311 UP000007266 UP000053097 UP000008237 UP000078541 UP000078542 UP000279307 UP000078492 UP000075809 UP000002358 UP000078540 UP000215335 UP000245037 UP000053105 UP000053201 UP000001554 UP000085678 UP000275408 UP000001593 UP000225706 UP000030746 UP000005408 UP000242188 UP000018721 UP000018958 UP000028582 UP000053236 UP000053864 UP000052943 UP000018817 UP000018948 UP000054423 UP000054532 UP000087266 UP000253843 UP000009022 UP000233060 UP000193380 UP000192224 UP000261440 UP000006643 UP000233140 UP000233120 UP000233080 UP000233180 UP000233100 UP000028761 UP000265515 UP000029965 UP000245341 UP000018468 UP000245340
PRIDE
Pfam
PF13870 DUF4201
Interpro
IPR025254
DUF4201
ProteinModelPortal
H9J170
A0A2A4JNF3
A0A194R507
A0A194PUU4
A0A2H1X1D1
A0A212FD22
+ More
A0A2A4JM89 A0A2H1WHX7 A0A0L7L9J8 A0A1W4XKN5 E2ASG8 A0A139WHB0 A0A026W8H0 E2BY35 E9IJW5 A0A195F7D1 A0A195D0B3 A0A3L8DI89 A0A195D9X7 A0A151WYE3 K7IUY2 A0A195B900 A0A232EUZ1 A0A2P8XL57 A0A0M8ZQS1 A0A0L0HK43 C3ZBE2 A0A1S3J674 A0A3M6UNB4 A7SJZ5 A0A2B4T1I5 V3YXT5 D2A2E0 A0A1B6DVC6 K1Q2F8 T2M5P4 A0A1B6LMP6 A0A210QH24 V9FKP8 W2XGN2 A0A081AMJ1 W2H7H4 A0A0W8D8M9 W2QGY5 W2ZPW0 W2LK35 A0A1S3MM96 A0A369SJG4 B3RVE1 D2A2D9 A0A2K5N0Z3 A0A060VXI0 A0A1W4Z333 A0A3B4C167 D0NZ21 A0A2K5Z423 A0A098M1B5 A0A2K6B738 A0A2K5K9W1 A0A2K6M7P5 A0A2K5UQZ9 A0A096NJ05 H9F3K1 A0A1S3MME4 A0A388KSS2 A0A0D9QXK7 A0A2U3XN24 W5M7M5 A0A2U3VLF9
A0A2A4JM89 A0A2H1WHX7 A0A0L7L9J8 A0A1W4XKN5 E2ASG8 A0A139WHB0 A0A026W8H0 E2BY35 E9IJW5 A0A195F7D1 A0A195D0B3 A0A3L8DI89 A0A195D9X7 A0A151WYE3 K7IUY2 A0A195B900 A0A232EUZ1 A0A2P8XL57 A0A0M8ZQS1 A0A0L0HK43 C3ZBE2 A0A1S3J674 A0A3M6UNB4 A7SJZ5 A0A2B4T1I5 V3YXT5 D2A2E0 A0A1B6DVC6 K1Q2F8 T2M5P4 A0A1B6LMP6 A0A210QH24 V9FKP8 W2XGN2 A0A081AMJ1 W2H7H4 A0A0W8D8M9 W2QGY5 W2ZPW0 W2LK35 A0A1S3MM96 A0A369SJG4 B3RVE1 D2A2D9 A0A2K5N0Z3 A0A060VXI0 A0A1W4Z333 A0A3B4C167 D0NZ21 A0A2K5Z423 A0A098M1B5 A0A2K6B738 A0A2K5K9W1 A0A2K6M7P5 A0A2K5UQZ9 A0A096NJ05 H9F3K1 A0A1S3MME4 A0A388KSS2 A0A0D9QXK7 A0A2U3XN24 W5M7M5 A0A2U3VLF9
Ontologies
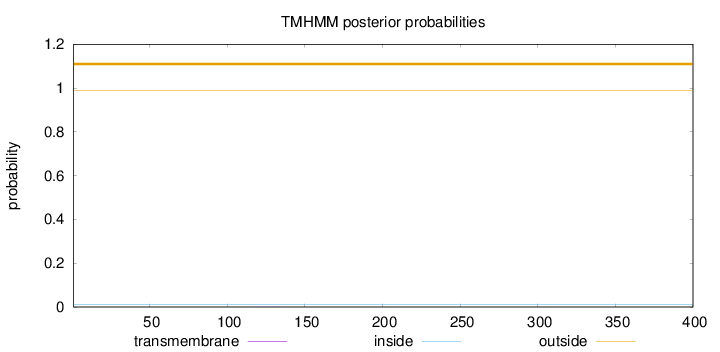
Topology
Length:
400
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00239
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01105
outside
1 - 400
Population Genetic Test Statistics
Pi
231.754458
Theta
206.981055
Tajima's D
0.376792
CLR
0
CSRT
0.472276386180691
Interpretation
Uncertain
