Pre Gene Modal
BGIBMGA003238
Annotation
PREDICTED:_probable_NADH_dehydrogenase_[ubiquinone]_1_alpha_subcomplex_subunit_12_[Amyelois_transitella]
Full name
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12
+ More
Serine/threonine-protein phosphatase 2A activator
Serine/threonine-protein phosphatase 2A activator
Alternative Name
Phosphotyrosyl phosphatase activator
Location in the cell
Extracellular Reliability : 1.303 Mitochondrial Reliability : 1.664
Sequence
CDS
ATGTCTCTGGGAAAGTTTTTAGCCCTCGACAAGCTAAGCAATTTTTTTAACGTAATTCGTCAGAACGGTGGTATAAGAGCCTCGTTGTATAAACTATACAGGCAGGACGATGTAAAGGACGGTGTTCTAGTTGGAGAAGACAAATATGGTAACAAATACTATGAGAACCCACGTTTCTTTTACAGCAGGAACAGATGGGTCGAGTACTCGGACAAGTATTACTTGAACTATGATGGAAGTCAGGTTCCCGCTGAATGGTTTGGTTGGCTACACTACAAAACCGATTTGCCTCCGCATCAGGATCCGAGTCGTCCCCACTACAAGTGGATGGCGGACCACACCGAAAATCTTTCTGGCACGACCGCCCAATATGTGCCGTACAGTACGACCCGACCCAAAGTGGAAGCCTGGGAGCCGAAACGCAAAGACAGTGTTTAA
Protein
MSLGKFLALDKLSNFFNVIRQNGGIRASLYKLYRQDDVKDGVLVGEDKYGNKYYENPRFFYSRNRWVEYSDKYYLNYDGSQVPAEWFGWLHYKTDLPPHQDPSRPHYKWMADHTENLSGTTAQYVPYSTTRPKVEAWEPKRKDSV
Summary
Description
Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I), that is believed not to be involved in catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone.
PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides.
PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides.
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Subunit
Complex I is composed of 45 different subunits.
Similarity
Belongs to the complex I NDUFA12 subunit family.
Belongs to the PTPA-type PPIase family.
Belongs to the PTPA-type PPIase family.
Uniprot
H9J151
A0A2W1BR08
A0A2A4JNC4
A0A2H1W2E6
A0A194QZR7
A0A194PT81
+ More
S4PTA1 A0A2R7W1X2 A0A067QUE7 A0A212FDH1 A0A0A9Z3S6 A0A146LY30 A0A1B0CW92 A0A0K8T375 A0A069DW08 A0A0P4VQY3 T1HN69 A0A146LNP5 A0A146LEA0 A0A2J7RA20 A0A224Y0T7 R4G4M7 A0A1B0DHU8 A0A0V0G8X9 D2A6F4 A0A182MD65 Q17M02 A0A182SL98 A0A023EGK7 A0A182RK31 A0A182Q7I9 A0A182NGR0 A0A084VSM7 A0A182V659 A0A1W4XQK1 A0A1Y9IW05 A0A182K1C7 A0A182X5U8 A0A1C7CZD0 A0A182U0R9 Q7Q8F5 A0A182IBI1 A0A1L8E0M2 W5J236 A0A1L8EHE4 A0A1I8PRP1 T1PEY0 A0A1J1IM65 A0A182P1M1 A0A2M4AEJ5 A0A2M3Z4W2 A0A1B6LZL4 A0A1I8MJ35 A0A2M4C2M4 W8C1Q7 A0A182F371 A0A034VWA4 A0A0C9RGY5 A0A0A1X7F9 A0A0L0CMB9 A0A0K8UDI9 A0A1A9XER4 A0A1B0C0P0 A0A1B0FH65 A0A1Q3FNF6 V5GT38 A0A1A9WKK0 B0XE21 A0A1A9V895 A0A1A9Z7X2 A0A026WT41 A0A0K8TTE2 A0A182JE99 A0A0L7RKB4 A0A195F279 U5ETD9 K7INC9 A0A158NG18 A0A1Y1L422 A0A151I1L3 B4N0E2 A0A0L7KT39 A0A2A3EHQ3 E9J5Y5 A0A195E806 A0A3B0KDY7 J3JX62 Q29KY1 B4GSM5 F4X6K1 B4KL35 A0A232EGE9 A0A336MVB2 A0A151ILK7 A0A1B6CJ33 B4JDB8 Q9VQD7 A0A151XB82 A0A0M4E7Z1 A0A0M3QU72
S4PTA1 A0A2R7W1X2 A0A067QUE7 A0A212FDH1 A0A0A9Z3S6 A0A146LY30 A0A1B0CW92 A0A0K8T375 A0A069DW08 A0A0P4VQY3 T1HN69 A0A146LNP5 A0A146LEA0 A0A2J7RA20 A0A224Y0T7 R4G4M7 A0A1B0DHU8 A0A0V0G8X9 D2A6F4 A0A182MD65 Q17M02 A0A182SL98 A0A023EGK7 A0A182RK31 A0A182Q7I9 A0A182NGR0 A0A084VSM7 A0A182V659 A0A1W4XQK1 A0A1Y9IW05 A0A182K1C7 A0A182X5U8 A0A1C7CZD0 A0A182U0R9 Q7Q8F5 A0A182IBI1 A0A1L8E0M2 W5J236 A0A1L8EHE4 A0A1I8PRP1 T1PEY0 A0A1J1IM65 A0A182P1M1 A0A2M4AEJ5 A0A2M3Z4W2 A0A1B6LZL4 A0A1I8MJ35 A0A2M4C2M4 W8C1Q7 A0A182F371 A0A034VWA4 A0A0C9RGY5 A0A0A1X7F9 A0A0L0CMB9 A0A0K8UDI9 A0A1A9XER4 A0A1B0C0P0 A0A1B0FH65 A0A1Q3FNF6 V5GT38 A0A1A9WKK0 B0XE21 A0A1A9V895 A0A1A9Z7X2 A0A026WT41 A0A0K8TTE2 A0A182JE99 A0A0L7RKB4 A0A195F279 U5ETD9 K7INC9 A0A158NG18 A0A1Y1L422 A0A151I1L3 B4N0E2 A0A0L7KT39 A0A2A3EHQ3 E9J5Y5 A0A195E806 A0A3B0KDY7 J3JX62 Q29KY1 B4GSM5 F4X6K1 B4KL35 A0A232EGE9 A0A336MVB2 A0A151ILK7 A0A1B6CJ33 B4JDB8 Q9VQD7 A0A151XB82 A0A0M4E7Z1 A0A0M3QU72
EC Number
5.2.1.8
Pubmed
19121390
28756777
26354079
23622113
24845553
22118469
+ More
25401762 26823975 26334808 27129103 18362917 19820115 17510324 24945155 26483478 24438588 20966253 12364791 14747013 17210077 20920257 23761445 25315136 24495485 25348373 25830018 26108605 24508170 26369729 20075255 21347285 28004739 17994087 26227816 21282665 22516182 23537049 15632085 21719571 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
25401762 26823975 26334808 27129103 18362917 19820115 17510324 24945155 26483478 24438588 20966253 12364791 14747013 17210077 20920257 23761445 25315136 24495485 25348373 25830018 26108605 24508170 26369729 20075255 21347285 28004739 17994087 26227816 21282665 22516182 23537049 15632085 21719571 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01031374
BABH01031375
KZ150065
PZC74173.1
NWSH01001044
PCG72943.1
+ More
ODYU01005900 SOQ47265.1 KQ460890 KPJ10947.1 KQ459593 KPI96522.1 GAIX01012398 JAA80162.1 KK854197 PTY12940.1 KK852999 KDR12671.1 AGBW02009033 OWR51815.1 GBHO01005606 JAG37998.1 GDHC01006754 JAQ11875.1 AJWK01032044 GBRD01005975 JAG59846.1 GBGD01003390 JAC85499.1 GDKW01001677 JAI54918.1 ACPB03020358 GDHC01009045 JAQ09584.1 GDHC01012680 JAQ05949.1 NEVH01006580 PNF37688.1 GFTR01002387 JAW14039.1 GAHY01001225 JAA76285.1 AJVK01034172 GECL01002285 JAP03839.1 KQ971347 EFA05745.1 AXCM01005884 CH477209 EAT47742.1 JXUM01151927 GAPW01005659 KQ571331 JAC07939.1 KXJ68202.1 AXCN02002079 ATLV01016091 KE525057 KFB40971.1 AAAB01008944 EAA10156.3 APCN01000703 GFDF01001888 JAV12196.1 ADMH02002210 ETN57796.1 GFDG01000734 JAV18065.1 KA647352 AFP61981.1 CVRI01000055 CRL01315.1 GGFK01005821 MBW39142.1 GGFM01002784 MBW23535.1 GEBQ01010850 JAT29127.1 GGFJ01010421 MBW59562.1 GAMC01000808 GAMC01000807 JAC05748.1 GAKP01012827 JAC46125.1 GBYB01012442 GBYB01012443 JAG82209.1 JAG82210.1 GBXI01007215 JAD07077.1 JRES01000186 KNC33493.1 GDHF01027610 GDHF01000211 JAI24704.1 JAI52103.1 JXJN01023698 CCAG010002037 GFDL01005981 JAV29064.1 GALX01003659 JAB64807.1 DS232798 EDS45793.1 KK107109 EZA59103.1 GDAI01000172 JAI17431.1 KQ414578 KOC71181.1 KQ981864 KYN34194.1 GANO01002818 JAB57053.1 ADTU01014889 GEZM01065539 JAV68432.1 KQ976580 KYM79897.1 CH963920 EDW77555.1 JTDY01006234 KOB66179.1 KZ288253 PBC30994.1 GL768221 EFZ11718.1 KQ979568 KYN20954.1 OUUW01000006 SPP81878.1 APGK01046266 BT127830 KB741050 KB631815 AEE62792.1 ENN74480.1 ERL86489.1 CH379061 EAL33043.1 CH479189 EDW25384.1 GL888818 EGI57961.1 CH933807 EDW12785.1 NNAY01004785 OXU17414.1 UFQS01000326 UFQS01002785 UFQT01000326 UFQT01002785 SSX02904.1 SSX34060.1 KQ977104 KYN05767.1 GEDC01023792 GEDC01016160 JAS13506.1 JAS21138.1 CH916368 EDW03291.1 AE014134 AAF51238.1 ACL82977.1 KQ982335 KYQ57604.1 CP012523 ALC38038.1 ALC40129.1
ODYU01005900 SOQ47265.1 KQ460890 KPJ10947.1 KQ459593 KPI96522.1 GAIX01012398 JAA80162.1 KK854197 PTY12940.1 KK852999 KDR12671.1 AGBW02009033 OWR51815.1 GBHO01005606 JAG37998.1 GDHC01006754 JAQ11875.1 AJWK01032044 GBRD01005975 JAG59846.1 GBGD01003390 JAC85499.1 GDKW01001677 JAI54918.1 ACPB03020358 GDHC01009045 JAQ09584.1 GDHC01012680 JAQ05949.1 NEVH01006580 PNF37688.1 GFTR01002387 JAW14039.1 GAHY01001225 JAA76285.1 AJVK01034172 GECL01002285 JAP03839.1 KQ971347 EFA05745.1 AXCM01005884 CH477209 EAT47742.1 JXUM01151927 GAPW01005659 KQ571331 JAC07939.1 KXJ68202.1 AXCN02002079 ATLV01016091 KE525057 KFB40971.1 AAAB01008944 EAA10156.3 APCN01000703 GFDF01001888 JAV12196.1 ADMH02002210 ETN57796.1 GFDG01000734 JAV18065.1 KA647352 AFP61981.1 CVRI01000055 CRL01315.1 GGFK01005821 MBW39142.1 GGFM01002784 MBW23535.1 GEBQ01010850 JAT29127.1 GGFJ01010421 MBW59562.1 GAMC01000808 GAMC01000807 JAC05748.1 GAKP01012827 JAC46125.1 GBYB01012442 GBYB01012443 JAG82209.1 JAG82210.1 GBXI01007215 JAD07077.1 JRES01000186 KNC33493.1 GDHF01027610 GDHF01000211 JAI24704.1 JAI52103.1 JXJN01023698 CCAG010002037 GFDL01005981 JAV29064.1 GALX01003659 JAB64807.1 DS232798 EDS45793.1 KK107109 EZA59103.1 GDAI01000172 JAI17431.1 KQ414578 KOC71181.1 KQ981864 KYN34194.1 GANO01002818 JAB57053.1 ADTU01014889 GEZM01065539 JAV68432.1 KQ976580 KYM79897.1 CH963920 EDW77555.1 JTDY01006234 KOB66179.1 KZ288253 PBC30994.1 GL768221 EFZ11718.1 KQ979568 KYN20954.1 OUUW01000006 SPP81878.1 APGK01046266 BT127830 KB741050 KB631815 AEE62792.1 ENN74480.1 ERL86489.1 CH379061 EAL33043.1 CH479189 EDW25384.1 GL888818 EGI57961.1 CH933807 EDW12785.1 NNAY01004785 OXU17414.1 UFQS01000326 UFQS01002785 UFQT01000326 UFQT01002785 SSX02904.1 SSX34060.1 KQ977104 KYN05767.1 GEDC01023792 GEDC01016160 JAS13506.1 JAS21138.1 CH916368 EDW03291.1 AE014134 AAF51238.1 ACL82977.1 KQ982335 KYQ57604.1 CP012523 ALC38038.1 ALC40129.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000027135
UP000007151
+ More
UP000092461 UP000015103 UP000235965 UP000092462 UP000007266 UP000075883 UP000008820 UP000075901 UP000069940 UP000249989 UP000075900 UP000075886 UP000075884 UP000030765 UP000075903 UP000192223 UP000075920 UP000075881 UP000076407 UP000075882 UP000075902 UP000007062 UP000075840 UP000000673 UP000095300 UP000183832 UP000075885 UP000095301 UP000069272 UP000037069 UP000092443 UP000092460 UP000092444 UP000091820 UP000002320 UP000078200 UP000092445 UP000053097 UP000075880 UP000053825 UP000078541 UP000002358 UP000005205 UP000078540 UP000007798 UP000037510 UP000242457 UP000078492 UP000268350 UP000019118 UP000030742 UP000001819 UP000008744 UP000007755 UP000009192 UP000215335 UP000078542 UP000001070 UP000000803 UP000075809 UP000092553
UP000092461 UP000015103 UP000235965 UP000092462 UP000007266 UP000075883 UP000008820 UP000075901 UP000069940 UP000249989 UP000075900 UP000075886 UP000075884 UP000030765 UP000075903 UP000192223 UP000075920 UP000075881 UP000076407 UP000075882 UP000075902 UP000007062 UP000075840 UP000000673 UP000095300 UP000183832 UP000075885 UP000095301 UP000069272 UP000037069 UP000092443 UP000092460 UP000092444 UP000091820 UP000002320 UP000078200 UP000092445 UP000053097 UP000075880 UP000053825 UP000078541 UP000002358 UP000005205 UP000078540 UP000007798 UP000037510 UP000242457 UP000078492 UP000268350 UP000019118 UP000030742 UP000001819 UP000008744 UP000007755 UP000009192 UP000215335 UP000078542 UP000001070 UP000000803 UP000075809 UP000092553
SUPFAM
SSF140984
SSF140984
CDD
ProteinModelPortal
H9J151
A0A2W1BR08
A0A2A4JNC4
A0A2H1W2E6
A0A194QZR7
A0A194PT81
+ More
S4PTA1 A0A2R7W1X2 A0A067QUE7 A0A212FDH1 A0A0A9Z3S6 A0A146LY30 A0A1B0CW92 A0A0K8T375 A0A069DW08 A0A0P4VQY3 T1HN69 A0A146LNP5 A0A146LEA0 A0A2J7RA20 A0A224Y0T7 R4G4M7 A0A1B0DHU8 A0A0V0G8X9 D2A6F4 A0A182MD65 Q17M02 A0A182SL98 A0A023EGK7 A0A182RK31 A0A182Q7I9 A0A182NGR0 A0A084VSM7 A0A182V659 A0A1W4XQK1 A0A1Y9IW05 A0A182K1C7 A0A182X5U8 A0A1C7CZD0 A0A182U0R9 Q7Q8F5 A0A182IBI1 A0A1L8E0M2 W5J236 A0A1L8EHE4 A0A1I8PRP1 T1PEY0 A0A1J1IM65 A0A182P1M1 A0A2M4AEJ5 A0A2M3Z4W2 A0A1B6LZL4 A0A1I8MJ35 A0A2M4C2M4 W8C1Q7 A0A182F371 A0A034VWA4 A0A0C9RGY5 A0A0A1X7F9 A0A0L0CMB9 A0A0K8UDI9 A0A1A9XER4 A0A1B0C0P0 A0A1B0FH65 A0A1Q3FNF6 V5GT38 A0A1A9WKK0 B0XE21 A0A1A9V895 A0A1A9Z7X2 A0A026WT41 A0A0K8TTE2 A0A182JE99 A0A0L7RKB4 A0A195F279 U5ETD9 K7INC9 A0A158NG18 A0A1Y1L422 A0A151I1L3 B4N0E2 A0A0L7KT39 A0A2A3EHQ3 E9J5Y5 A0A195E806 A0A3B0KDY7 J3JX62 Q29KY1 B4GSM5 F4X6K1 B4KL35 A0A232EGE9 A0A336MVB2 A0A151ILK7 A0A1B6CJ33 B4JDB8 Q9VQD7 A0A151XB82 A0A0M4E7Z1 A0A0M3QU72
S4PTA1 A0A2R7W1X2 A0A067QUE7 A0A212FDH1 A0A0A9Z3S6 A0A146LY30 A0A1B0CW92 A0A0K8T375 A0A069DW08 A0A0P4VQY3 T1HN69 A0A146LNP5 A0A146LEA0 A0A2J7RA20 A0A224Y0T7 R4G4M7 A0A1B0DHU8 A0A0V0G8X9 D2A6F4 A0A182MD65 Q17M02 A0A182SL98 A0A023EGK7 A0A182RK31 A0A182Q7I9 A0A182NGR0 A0A084VSM7 A0A182V659 A0A1W4XQK1 A0A1Y9IW05 A0A182K1C7 A0A182X5U8 A0A1C7CZD0 A0A182U0R9 Q7Q8F5 A0A182IBI1 A0A1L8E0M2 W5J236 A0A1L8EHE4 A0A1I8PRP1 T1PEY0 A0A1J1IM65 A0A182P1M1 A0A2M4AEJ5 A0A2M3Z4W2 A0A1B6LZL4 A0A1I8MJ35 A0A2M4C2M4 W8C1Q7 A0A182F371 A0A034VWA4 A0A0C9RGY5 A0A0A1X7F9 A0A0L0CMB9 A0A0K8UDI9 A0A1A9XER4 A0A1B0C0P0 A0A1B0FH65 A0A1Q3FNF6 V5GT38 A0A1A9WKK0 B0XE21 A0A1A9V895 A0A1A9Z7X2 A0A026WT41 A0A0K8TTE2 A0A182JE99 A0A0L7RKB4 A0A195F279 U5ETD9 K7INC9 A0A158NG18 A0A1Y1L422 A0A151I1L3 B4N0E2 A0A0L7KT39 A0A2A3EHQ3 E9J5Y5 A0A195E806 A0A3B0KDY7 J3JX62 Q29KY1 B4GSM5 F4X6K1 B4KL35 A0A232EGE9 A0A336MVB2 A0A151ILK7 A0A1B6CJ33 B4JDB8 Q9VQD7 A0A151XB82 A0A0M4E7Z1 A0A0M3QU72
PDB
5LDX
E-value=6.37851e-26,
Score=285
Ontologies
PATHWAY
GO
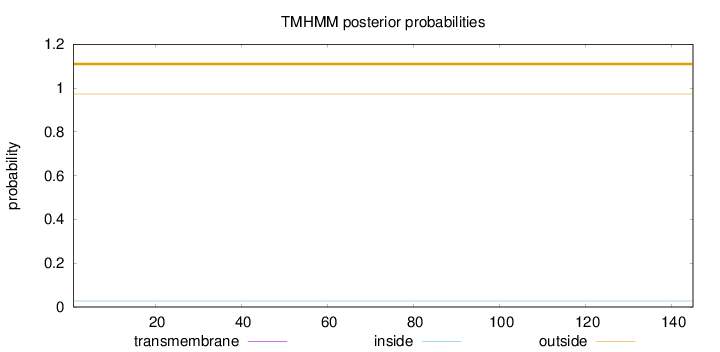
Topology
Subcellular location
Mitochondrion inner membrane
Cytoplasm
Cytoplasm
Length:
145
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000670000000000001
Exp number, first 60 AAs:
0.00049
Total prob of N-in:
0.02723
outside
1 - 145
Population Genetic Test Statistics
Pi
160.509603
Theta
172.721597
Tajima's D
-0.957378
CLR
0.100676
CSRT
0.149742512874356
Interpretation
Uncertain
