Gene
KWMTBOMO00723 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003258
Annotation
pyruvate_dehydrogenase_kinase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.084 Mitochondrial Reliability : 1.742
Sequence
CDS
ATGACCCGCGACTCTTGGCGCGTCAATACAGGTTTACGCGATACGGAAGGTCTGAACGCTTGCGAGAGCAAATCATTCACGTTCCTAAAGAAGGAGCTACCAGTCAGGCTCGCGAACATAATGAAGGAGATAGCGTTACTCCCTGAGAACCTACTTCGAATGCCTTCGGTGGGTTTAGTCAACCAATGGTACGAACGCTCATTCGAGGAAATAACACGCTTTGAACAGATGGAACCAGATCCTCCGACGCTGACCCAGTTCTGCGAACGTCTTGTCCACATAAGGAACCGGCACGCTGACGTCGTTCAGACGATGGCCCAGGGTGTGCTAGAGCTGAAAGAGTCTCACGAAGTTGATCCCGGAACGGAGAACTCCATACAATACTTCCTGGACAGATTCTACATGAGTCGGATATCTATTCGGATGCTTATTAACCAGCACACTCTGTTGTTTGGCGAGCAGCTTGGCGGCAAGCAGGCGTCGGTCAATGGGATCGGCAATGGTGGCCGTCACATCGGCTCCATAGACCCGGCCTGCGACGTCGTGGCCGTCGTTAGAGACGCCTACGAGAACGCAAGGTTCCTCTGCGATAGATACTACCTAGCCTCACCAGAGTTGGAGGTACTGCAGGACGGAGTGTCGAGTCTGCGCCCGATGCCCATAGTGTATGTGCCCTCGCACCTCTACCACATGCTGTTTGAGCTATTCAAGAATGCCATGAGGGCCGTCATGGAGAACCACGGCACCGCCCCACCGCCGATACAAGTTAACCTCGTCAACGGGAAAGAGGACATATCCGTTAAGATGTCTGATCGCGGCGGCGGCATCCCGCGCAGCGTGTCGGAGCTTCTGTTCAAGTACATGTACAGCACGGCGCCGCAGCCCTCCAAGTCCGACTCGCACACGGTGCCGCTAGCGGGTTACGGATACGGGCTGCCCATCTCTCGCCTGTACGCGCGGTACTTCCACGGCGACCTGGTGCTGGTATCGTGCGAGGGTTACGGCACGGACGCCGTCATCTACCTCAAGGCTCTAACGAACGAAGCCAACGAACTACTGCCGGTTTTCAATAAGACGTCTTCGAAGTTCTACCGCCCCTCGTCCCAGCTGGCGGACTGGTCCGGCACCCTCAACTCCTCGAATCAGTGCAGGAGGGAGAAAGTGTCTCCGCTCTCATCACCGATATCGCACAGCCTATAG
Protein
MTRDSWRVNTGLRDTEGLNACESKSFTFLKKELPVRLANIMKEIALLPENLLRMPSVGLVNQWYERSFEEITRFEQMEPDPPTLTQFCERLVHIRNRHADVVQTMAQGVLELKESHEVDPGTENSIQYFLDRFYMSRISIRMLINQHTLLFGEQLGGKQASVNGIGNGGRHIGSIDPACDVVAVVRDAYENARFLCDRYYLASPELEVLQDGVSSLRPMPIVYVPSHLYHMLFELFKNAMRAVMENHGTAPPPIQVNLVNGKEDISVKMSDRGGGIPRSVSELLFKYMYSTAPQPSKSDSHTVPLAGYGYGLPISRLYARYFHGDLVLVSCEGYGTDAVIYLKALTNEANELLPVFNKTSSKFYRPSSQLADWSGTLNSSNQCRREKVSPLSSPISHSL
Summary
Uniprot
B0LL83
H9J171
A0A194R0U4
A0A2H1WE60
A0A2A4K732
A0A194PSQ1
+ More
A0A212F768 A0A2J7RKE6 A0A1B6DQJ9 D6WRE8 A0A026WJV4 A0A1B6FGS4 A0A195FPH5 A0A1B6HGU3 A0A1S3DCA9 A0A151JR10 A0A3L8D9Y1 A0A195CU95 A0A195BMD8 A0A158NTW3 A0A1Y1LGX4 K7IQX1 A0A151WLN9 N6TUF7 A0A1B6KU71 A0A336LWG4 V5I8M0 A0A154P5I6 V5G1Y8 A0A182RAE8 A0A182M8M2 A0A2S2N6D1 A0A182PPU4 A0A069DU55 A0A2M4BQY6 A0A232F4Y0 A0A182JCK9 A0A2M4AGY1 A0A182F8V7 Q172I0 A0A2M4CLL3 A0A2M4CLF0 A0A2H8TVL0 A0A088AA34 A0A1S4H3I4 A0A023ETR1 A0A023F4M8 A0A2A3EJB4 Q172I2 Q172I1 A0A2M4BPM9 A0A2M4BQH6 U5EZ53 W5JFI0 A0A2M3Z804 A0A2M4AGN4 A0A1S4H3I3 A0A2M3ZHY9 A0A1Q3FYE2 A0A0P4VPN9 A0A0L7R6L0 A0A1S4K0W2 B0X1X9 A0A182VSS6 A0A224XDQ5 A0A0K8TM15 A0A1W4XGV8 E0W1B8 A0A182I0W4 T1E232 A0A0V0G6I1 A0A0L0CA08 A0A182XJD9 T1PLG5 T1E234 A0A182TQF8 A0A182KJX7 A0A0A1X0B8 T1DI51 W8B2X5 T1PLE9 A0A1I8NC03 J9K332 A0A0K8V7Y1 A0A1B0C5U9 A0A1J1IH56 A0A1A9YS53 A0A0A9X1W4 A0A034VHP2 D3TM48 A0A1A9WQJ8 A0A1A9VPQ3 A0A1J1IKH7 E1ZXR8 A0A1I8Q6Y3 B3ME72 A0A0N8NZZ2 A0A1A9ZB64 B4MRI1 B4NWG0
A0A212F768 A0A2J7RKE6 A0A1B6DQJ9 D6WRE8 A0A026WJV4 A0A1B6FGS4 A0A195FPH5 A0A1B6HGU3 A0A1S3DCA9 A0A151JR10 A0A3L8D9Y1 A0A195CU95 A0A195BMD8 A0A158NTW3 A0A1Y1LGX4 K7IQX1 A0A151WLN9 N6TUF7 A0A1B6KU71 A0A336LWG4 V5I8M0 A0A154P5I6 V5G1Y8 A0A182RAE8 A0A182M8M2 A0A2S2N6D1 A0A182PPU4 A0A069DU55 A0A2M4BQY6 A0A232F4Y0 A0A182JCK9 A0A2M4AGY1 A0A182F8V7 Q172I0 A0A2M4CLL3 A0A2M4CLF0 A0A2H8TVL0 A0A088AA34 A0A1S4H3I4 A0A023ETR1 A0A023F4M8 A0A2A3EJB4 Q172I2 Q172I1 A0A2M4BPM9 A0A2M4BQH6 U5EZ53 W5JFI0 A0A2M3Z804 A0A2M4AGN4 A0A1S4H3I3 A0A2M3ZHY9 A0A1Q3FYE2 A0A0P4VPN9 A0A0L7R6L0 A0A1S4K0W2 B0X1X9 A0A182VSS6 A0A224XDQ5 A0A0K8TM15 A0A1W4XGV8 E0W1B8 A0A182I0W4 T1E232 A0A0V0G6I1 A0A0L0CA08 A0A182XJD9 T1PLG5 T1E234 A0A182TQF8 A0A182KJX7 A0A0A1X0B8 T1DI51 W8B2X5 T1PLE9 A0A1I8NC03 J9K332 A0A0K8V7Y1 A0A1B0C5U9 A0A1J1IH56 A0A1A9YS53 A0A0A9X1W4 A0A034VHP2 D3TM48 A0A1A9WQJ8 A0A1A9VPQ3 A0A1J1IKH7 E1ZXR8 A0A1I8Q6Y3 B3ME72 A0A0N8NZZ2 A0A1A9ZB64 B4MRI1 B4NWG0
Pubmed
20197069
19121390
26354079
22118469
18362917
19820115
+ More
24508170 30249741 21347285 28004739 20075255 23537049 26334808 28648823 17510324 12364791 24945155 26483478 25474469 20920257 23761445 27129103 26369729 20566863 24330624 26108605 20966253 25830018 24495485 25315136 25401762 25348373 20353571 20798317 17994087 18057021 17550304
24508170 30249741 21347285 28004739 20075255 23537049 26334808 28648823 17510324 12364791 24945155 26483478 25474469 20920257 23761445 27129103 26369729 20566863 24330624 26108605 20966253 25830018 24495485 25315136 25401762 25348373 20353571 20798317 17994087 18057021 17550304
EMBL
EU249373
ABY76060.1
BABH01031383
KQ460890
KPJ10870.1
ODYU01008076
+ More
SOQ51370.1 NWSH01000062 PCG80047.1 KQ459593 KPI96461.1 AGBW02009910 OWR49586.1 NEVH01002981 PNF41302.1 GEDC01029767 GEDC01009348 JAS07531.1 JAS27950.1 KQ971351 EFA06557.2 KK107167 EZA56243.1 GECZ01022868 GECZ01021935 GECZ01020361 JAS46901.1 JAS47834.1 JAS49408.1 KQ981382 KYN42301.1 GECU01033814 JAS73892.1 KQ978615 KYN29829.1 QOIP01000010 RLU17287.1 KQ977279 KYN04097.1 KQ976435 KYM87053.1 ADTU01026166 GEZM01057705 JAV72131.1 KQ982959 KYQ48753.1 APGK01054105 APGK01054106 KB741247 KB632281 ENN72016.1 ERL91354.1 GEBQ01024982 GEBQ01003801 JAT14995.1 JAT36176.1 UFQT01000186 SSX21311.1 GALX01004450 JAB64016.1 KQ434822 KZC07111.1 GALX01004451 JAB64015.1 AXCM01000163 GGMR01000030 MBY12649.1 GBGD01001662 JAC87227.1 GGFJ01006047 MBW55188.1 NNAY01000957 OXU25735.1 GGFK01006720 MBW40041.1 CH477436 EAT40947.1 GGFL01001991 MBW66169.1 GGFL01001992 MBW66170.1 GFXV01005787 MBW17592.1 AAAB01008980 JXUM01036156 JXUM01036157 JXUM01036158 GAPW01001794 KQ561085 JAC11804.1 KXJ79758.1 GBBI01002390 GEMB01002970 JAC16322.1 JAS00233.1 KZ288249 PBC31121.1 EAT40944.1 EAT40945.1 GGFJ01005821 MBW54962.1 GGFJ01006082 MBW55223.1 GANO01001414 JAB58457.1 ADMH02001363 ETN62831.1 GGFM01003905 MBW24656.1 GGFK01006618 MBW39939.1 GGFM01007381 MBW28132.1 GFDL01002441 JAV32604.1 GDKW01002253 JAI54342.1 KQ414646 KOC66483.1 DS232274 EDS38909.1 GFTR01005841 JAW10585.1 GDAI01002204 JAI15399.1 DS235867 EEB19424.1 APCN01005279 GALA01001104 JAA93748.1 GECL01002428 JAP03696.1 JRES01000678 KNC29263.1 KA646693 KA649580 KA649601 AFP64230.1 GALA01001099 JAA93753.1 GBXI01009533 JAD04759.1 GALA01001141 JAA93711.1 GAMC01015162 GAMC01015161 JAB91393.1 KA649600 KA649602 AFP64231.1 ABLF02013322 GDHF01017629 JAI34685.1 JXJN01012881 JXJN01026246 CVRI01000051 CRK99581.1 GBHO01032524 GBRD01016834 GBRD01008185 GBRD01008184 JAG11080.1 JAG48993.1 GAKP01017320 GAKP01017319 JAC41632.1 EZ422500 ADD18776.1 CRK99582.1 GL435087 EFN74040.1 CH902619 EDV35467.1 KPU75675.1 KPU75676.1 CH963850 EDW74720.1 CM000157 EDW89505.2
SOQ51370.1 NWSH01000062 PCG80047.1 KQ459593 KPI96461.1 AGBW02009910 OWR49586.1 NEVH01002981 PNF41302.1 GEDC01029767 GEDC01009348 JAS07531.1 JAS27950.1 KQ971351 EFA06557.2 KK107167 EZA56243.1 GECZ01022868 GECZ01021935 GECZ01020361 JAS46901.1 JAS47834.1 JAS49408.1 KQ981382 KYN42301.1 GECU01033814 JAS73892.1 KQ978615 KYN29829.1 QOIP01000010 RLU17287.1 KQ977279 KYN04097.1 KQ976435 KYM87053.1 ADTU01026166 GEZM01057705 JAV72131.1 KQ982959 KYQ48753.1 APGK01054105 APGK01054106 KB741247 KB632281 ENN72016.1 ERL91354.1 GEBQ01024982 GEBQ01003801 JAT14995.1 JAT36176.1 UFQT01000186 SSX21311.1 GALX01004450 JAB64016.1 KQ434822 KZC07111.1 GALX01004451 JAB64015.1 AXCM01000163 GGMR01000030 MBY12649.1 GBGD01001662 JAC87227.1 GGFJ01006047 MBW55188.1 NNAY01000957 OXU25735.1 GGFK01006720 MBW40041.1 CH477436 EAT40947.1 GGFL01001991 MBW66169.1 GGFL01001992 MBW66170.1 GFXV01005787 MBW17592.1 AAAB01008980 JXUM01036156 JXUM01036157 JXUM01036158 GAPW01001794 KQ561085 JAC11804.1 KXJ79758.1 GBBI01002390 GEMB01002970 JAC16322.1 JAS00233.1 KZ288249 PBC31121.1 EAT40944.1 EAT40945.1 GGFJ01005821 MBW54962.1 GGFJ01006082 MBW55223.1 GANO01001414 JAB58457.1 ADMH02001363 ETN62831.1 GGFM01003905 MBW24656.1 GGFK01006618 MBW39939.1 GGFM01007381 MBW28132.1 GFDL01002441 JAV32604.1 GDKW01002253 JAI54342.1 KQ414646 KOC66483.1 DS232274 EDS38909.1 GFTR01005841 JAW10585.1 GDAI01002204 JAI15399.1 DS235867 EEB19424.1 APCN01005279 GALA01001104 JAA93748.1 GECL01002428 JAP03696.1 JRES01000678 KNC29263.1 KA646693 KA649580 KA649601 AFP64230.1 GALA01001099 JAA93753.1 GBXI01009533 JAD04759.1 GALA01001141 JAA93711.1 GAMC01015162 GAMC01015161 JAB91393.1 KA649600 KA649602 AFP64231.1 ABLF02013322 GDHF01017629 JAI34685.1 JXJN01012881 JXJN01026246 CVRI01000051 CRK99581.1 GBHO01032524 GBRD01016834 GBRD01008185 GBRD01008184 JAG11080.1 JAG48993.1 GAKP01017320 GAKP01017319 JAC41632.1 EZ422500 ADD18776.1 CRK99582.1 GL435087 EFN74040.1 CH902619 EDV35467.1 KPU75675.1 KPU75676.1 CH963850 EDW74720.1 CM000157 EDW89505.2
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000007151
UP000235965
+ More
UP000007266 UP000053097 UP000078541 UP000079169 UP000078492 UP000279307 UP000078542 UP000078540 UP000005205 UP000002358 UP000075809 UP000019118 UP000030742 UP000076502 UP000075900 UP000075883 UP000075885 UP000215335 UP000075880 UP000069272 UP000008820 UP000005203 UP000069940 UP000249989 UP000242457 UP000000673 UP000053825 UP000002320 UP000075920 UP000192223 UP000009046 UP000075840 UP000037069 UP000076407 UP000075902 UP000075882 UP000095301 UP000007819 UP000092460 UP000183832 UP000092443 UP000091820 UP000078200 UP000000311 UP000095300 UP000007801 UP000092445 UP000007798 UP000002282
UP000007266 UP000053097 UP000078541 UP000079169 UP000078492 UP000279307 UP000078542 UP000078540 UP000005205 UP000002358 UP000075809 UP000019118 UP000030742 UP000076502 UP000075900 UP000075883 UP000075885 UP000215335 UP000075880 UP000069272 UP000008820 UP000005203 UP000069940 UP000249989 UP000242457 UP000000673 UP000053825 UP000002320 UP000075920 UP000192223 UP000009046 UP000075840 UP000037069 UP000076407 UP000075902 UP000075882 UP000095301 UP000007819 UP000092460 UP000183832 UP000092443 UP000091820 UP000078200 UP000000311 UP000095300 UP000007801 UP000092445 UP000007798 UP000002282
Interpro
Gene 3D
ProteinModelPortal
B0LL83
H9J171
A0A194R0U4
A0A2H1WE60
A0A2A4K732
A0A194PSQ1
+ More
A0A212F768 A0A2J7RKE6 A0A1B6DQJ9 D6WRE8 A0A026WJV4 A0A1B6FGS4 A0A195FPH5 A0A1B6HGU3 A0A1S3DCA9 A0A151JR10 A0A3L8D9Y1 A0A195CU95 A0A195BMD8 A0A158NTW3 A0A1Y1LGX4 K7IQX1 A0A151WLN9 N6TUF7 A0A1B6KU71 A0A336LWG4 V5I8M0 A0A154P5I6 V5G1Y8 A0A182RAE8 A0A182M8M2 A0A2S2N6D1 A0A182PPU4 A0A069DU55 A0A2M4BQY6 A0A232F4Y0 A0A182JCK9 A0A2M4AGY1 A0A182F8V7 Q172I0 A0A2M4CLL3 A0A2M4CLF0 A0A2H8TVL0 A0A088AA34 A0A1S4H3I4 A0A023ETR1 A0A023F4M8 A0A2A3EJB4 Q172I2 Q172I1 A0A2M4BPM9 A0A2M4BQH6 U5EZ53 W5JFI0 A0A2M3Z804 A0A2M4AGN4 A0A1S4H3I3 A0A2M3ZHY9 A0A1Q3FYE2 A0A0P4VPN9 A0A0L7R6L0 A0A1S4K0W2 B0X1X9 A0A182VSS6 A0A224XDQ5 A0A0K8TM15 A0A1W4XGV8 E0W1B8 A0A182I0W4 T1E232 A0A0V0G6I1 A0A0L0CA08 A0A182XJD9 T1PLG5 T1E234 A0A182TQF8 A0A182KJX7 A0A0A1X0B8 T1DI51 W8B2X5 T1PLE9 A0A1I8NC03 J9K332 A0A0K8V7Y1 A0A1B0C5U9 A0A1J1IH56 A0A1A9YS53 A0A0A9X1W4 A0A034VHP2 D3TM48 A0A1A9WQJ8 A0A1A9VPQ3 A0A1J1IKH7 E1ZXR8 A0A1I8Q6Y3 B3ME72 A0A0N8NZZ2 A0A1A9ZB64 B4MRI1 B4NWG0
A0A212F768 A0A2J7RKE6 A0A1B6DQJ9 D6WRE8 A0A026WJV4 A0A1B6FGS4 A0A195FPH5 A0A1B6HGU3 A0A1S3DCA9 A0A151JR10 A0A3L8D9Y1 A0A195CU95 A0A195BMD8 A0A158NTW3 A0A1Y1LGX4 K7IQX1 A0A151WLN9 N6TUF7 A0A1B6KU71 A0A336LWG4 V5I8M0 A0A154P5I6 V5G1Y8 A0A182RAE8 A0A182M8M2 A0A2S2N6D1 A0A182PPU4 A0A069DU55 A0A2M4BQY6 A0A232F4Y0 A0A182JCK9 A0A2M4AGY1 A0A182F8V7 Q172I0 A0A2M4CLL3 A0A2M4CLF0 A0A2H8TVL0 A0A088AA34 A0A1S4H3I4 A0A023ETR1 A0A023F4M8 A0A2A3EJB4 Q172I2 Q172I1 A0A2M4BPM9 A0A2M4BQH6 U5EZ53 W5JFI0 A0A2M3Z804 A0A2M4AGN4 A0A1S4H3I3 A0A2M3ZHY9 A0A1Q3FYE2 A0A0P4VPN9 A0A0L7R6L0 A0A1S4K0W2 B0X1X9 A0A182VSS6 A0A224XDQ5 A0A0K8TM15 A0A1W4XGV8 E0W1B8 A0A182I0W4 T1E232 A0A0V0G6I1 A0A0L0CA08 A0A182XJD9 T1PLG5 T1E234 A0A182TQF8 A0A182KJX7 A0A0A1X0B8 T1DI51 W8B2X5 T1PLE9 A0A1I8NC03 J9K332 A0A0K8V7Y1 A0A1B0C5U9 A0A1J1IH56 A0A1A9YS53 A0A0A9X1W4 A0A034VHP2 D3TM48 A0A1A9WQJ8 A0A1A9VPQ3 A0A1J1IKH7 E1ZXR8 A0A1I8Q6Y3 B3ME72 A0A0N8NZZ2 A0A1A9ZB64 B4MRI1 B4NWG0
PDB
2Q8H
E-value=7.90462e-118,
Score=1084
Ontologies
PANTHER
Topology
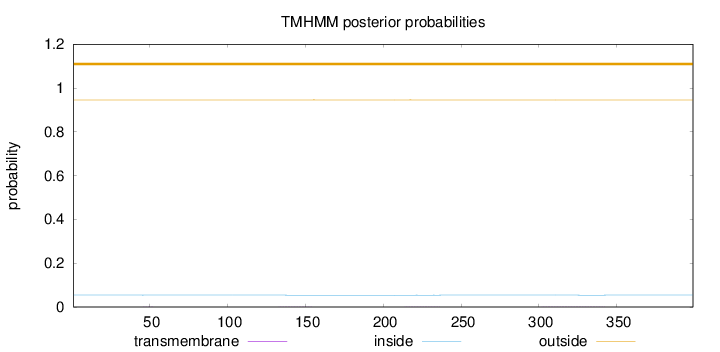
Length:
399
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02554
Exp number, first 60 AAs:
0.0023
Total prob of N-in:
0.05458
outside
1 - 399
Population Genetic Test Statistics
Pi
231.666651
Theta
150.92049
Tajima's D
2.008895
CLR
0.101695
CSRT
0.88415579221039
Interpretation
Uncertain
