Gene
KWMTBOMO00711 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003267
Annotation
PREDICTED:_protein_canopy_homolog_1_[Papilio_polytes]
Full name
Protein seele
Location in the cell
Nuclear Reliability : 2.38
Sequence
CDS
ATGAGATTGAATAGTTTAATTTTTGCATTTGTGATTTTATTCAGTGTTGTTTCAGCAAGAATCGACCCTAAAAACTTAAAATGTTTGGTTTGTCGGACGACGTTTGAGGAGCTTCACCAGGTGATAAAAAGCATAGATAAATGGAAAAGAGTCGATGTTGGCAACTTCAGAATGGACAAAGATGGCAACACAATGCAGCAGAAAGTGCCCGCTCATCGGTCTGCGGTCTACATTTCTGAACTTATTGATGAGATCTGCAAAAAGATGGATGACTACGTCCGCGTTTACTACAAGTCGACCGGTAAACTGACGATAATGCAGCTGATGACCAAGGAGGGCGGCATGAACCCTGAATTCTCGAAGACCAAATTCGTCACCGACGACGACCTGAACAAAAGCTTGGAGTATTACTGCGAACGTATGTTTGAAGACAACGAGGACGAGATCACGTCTTTGTACGTGAAGCGACCCGACGATGACGTCATGCCGGACGCGGAACGCGAGATATGCTTCAACCACGCCAAGTATTGCGAGGAGTGGATGTTACCCACTGAAGAGGACACGACTTGGACCAGAGAAATGGAGGAGGAGTATGTCAAGGTCCACGGGCCGGACCCGTACGGCTTTGGGGGCGTGCCGCCGCCCGCCCATCCCGACCTGTCCGAGGACTACGACGACACCGACCAACAGACCGACGACTTCGGCGACGACATCAGGGACGAGCTCTAG
Protein
MRLNSLIFAFVILFSVVSARIDPKNLKCLVCRTTFEELHQVIKSIDKWKRVDVGNFRMDKDGNTMQQKVPAHRSAVYISELIDEICKKMDDYVRVYYKSTGKLTIMQLMTKEGGMNPEFSKTKFVTDDDLNKSLEYYCERMFEDNEDEITSLYVKRPDDDVMPDAEREICFNHAKYCEEWMLPTEEDTTWTREMEEEYVKVHGPDPYGFGGVPPPAHPDLSEDYDDTDQQTDDFGDDIRDEL
Summary
Description
Involved in embryonic dorsal-ventral patterning which is generated by a series of serine protease processing events where gd processes snk which cleaves ea which then processes spz into the activating ligand for the Toll receptor. Required during this process for the secretion of ea from the developing embryo into the perivitelline space and for ea processing.
Similarity
Belongs to the canopy family.
Keywords
Complete proteome
Developmental protein
Disulfide bond
Endoplasmic reticulum
Reference proteome
Signal
Feature
chain Protein seele
Uniprot
A0A2H1VES3
A0A212ETX8
H9J180
A0A1E1WN67
A0A0N1I653
A0A088AH64
+ More
A0A2A3ETX6 A0A195E6J8 A0A151I932 A0A195EVC2 A0A158NR17 A0A195B0A9 E9ISN7 D6WRV5 A0A0M8ZYS0 A0A1Y1K0K3 A0A336MGW9 A0A2J7QF17 A0A0C9RKH5 A0A232F7L9 K7IYX5 A0A0K8TJB0 A0A026WNY7 A0A1L8DW48 A0A1L8DW36 A0A1L8DW46 A0A1L8DWL6 A0A0A9XIC9 A0A1S4FXN3 A0A0M4EHF4 A0A1B6HZD7 B4J5N5 Q1HR27 U4UR88 B4MQX2 B4LP39 N6UC79 T1J2X4 A0A2H8TFJ5 Q290T5 B4GB99 A0A023EJE0 Q16KG3 A0A0L0C7M6 A0A1I8MAC8 A0A2S2R826 A0A1B6F505 A0A3B0IZI8 A0A023ELJ2 A0A1L8EIG1 A0A182G7E1 A0A3L8DWU6 A0A2M3Z5D4 A0A224XYW7 A0A182F6A9 A0A1W4VDN5 B4KM38 B3N6J7 A0A2M4BZM3 A0A034WUZ1 A0A0K8VLW0 A0A2M4BZK8 B4NX39 A0A2M4A4D5 B4QI80 Q7JXF7 A0A2M4A491 B3MD24 A0A1Q3FCA7 W5JAU4 B0WLK3 B4HMB8 A0A0A1XHT4 A0A0J7L8K5 A0A1A9USS0 B4IMW9 A0A1A9YPB8 A0A1B0C6I0 E9H737
A0A2A3ETX6 A0A195E6J8 A0A151I932 A0A195EVC2 A0A158NR17 A0A195B0A9 E9ISN7 D6WRV5 A0A0M8ZYS0 A0A1Y1K0K3 A0A336MGW9 A0A2J7QF17 A0A0C9RKH5 A0A232F7L9 K7IYX5 A0A0K8TJB0 A0A026WNY7 A0A1L8DW48 A0A1L8DW36 A0A1L8DW46 A0A1L8DWL6 A0A0A9XIC9 A0A1S4FXN3 A0A0M4EHF4 A0A1B6HZD7 B4J5N5 Q1HR27 U4UR88 B4MQX2 B4LP39 N6UC79 T1J2X4 A0A2H8TFJ5 Q290T5 B4GB99 A0A023EJE0 Q16KG3 A0A0L0C7M6 A0A1I8MAC8 A0A2S2R826 A0A1B6F505 A0A3B0IZI8 A0A023ELJ2 A0A1L8EIG1 A0A182G7E1 A0A3L8DWU6 A0A2M3Z5D4 A0A224XYW7 A0A182F6A9 A0A1W4VDN5 B4KM38 B3N6J7 A0A2M4BZM3 A0A034WUZ1 A0A0K8VLW0 A0A2M4BZK8 B4NX39 A0A2M4A4D5 B4QI80 Q7JXF7 A0A2M4A491 B3MD24 A0A1Q3FCA7 W5JAU4 B0WLK3 B4HMB8 A0A0A1XHT4 A0A0J7L8K5 A0A1A9USS0 B4IMW9 A0A1A9YPB8 A0A1B0C6I0 E9H737
Pubmed
22118469
19121390
26354079
21347285
21282665
18362917
+ More
19820115 28004739 28648823 20075255 24508170 25401762 26823975 17994087 17204158 23537049 15632085 24945155 17510324 26108605 25315136 26483478 30249741 25348373 17550304 22936249 10731132 12537572 12537569 20970335 20920257 23761445 25830018 21292972
19820115 28004739 28648823 20075255 24508170 25401762 26823975 17994087 17204158 23537049 15632085 24945155 17510324 26108605 25315136 26483478 30249741 25348373 17550304 22936249 10731132 12537572 12537569 20970335 20920257 23761445 25830018 21292972
EMBL
ODYU01002169
SOQ39317.1
AGBW02012514
OWR44955.1
BABH01031435
GDQN01002758
+ More
JAT88296.1 KQ458499 KPJ05858.1 KZ288189 PBC34666.1 KQ979608 KYN20474.1 KQ978324 KYM95109.1 KQ981965 KYN31839.1 ADTU01023744 KQ976691 KYM77913.1 GL765434 EFZ16278.1 KQ971351 EFA06585.2 KQ435794 KOX73850.1 GEZM01101845 GEZM01101844 JAV52257.1 UFQT01001052 SSX28651.1 NEVH01015304 PNF27174.1 GBYB01008700 JAG78467.1 NNAY01000791 OXU26490.1 GBRD01000242 JAG65579.1 KK107139 EZA57730.1 GFDF01003426 JAV10658.1 GFDF01003427 JAV10657.1 GFDF01003428 JAV10656.1 GFDF01003429 JAV10655.1 GBHO01025051 GDHC01004043 JAG18553.1 JAQ14586.1 CP012524 ALC41985.1 GECU01027658 JAS80048.1 CH916367 EDW01811.1 DQ440267 ABF18300.1 KB632333 ERL92671.1 CH963849 EDW74511.1 CH940648 EDW62233.2 APGK01035263 KB740923 ENN78236.1 JH431812 GFXV01001059 GFXV01002379 MBW12864.1 MBW14184.1 CM000071 EAL25277.1 CH479181 EDW32201.1 GAPW01004533 JAC09065.1 CH477959 EAT34790.1 JRES01000780 KNC28378.1 GGMS01017004 MBY86207.1 GECZ01024437 JAS45332.1 OUUW01000001 SPP73625.1 GAPW01004480 GAPW01004479 JAC09119.1 GFDG01000400 JAV18399.1 JXUM01046464 KQ561478 KXJ78464.1 QOIP01000003 RLU24944.1 GGFM01002964 MBW23715.1 GFTR01003147 JAW13279.1 CH933808 EDW08702.2 CH954177 EDV58166.1 GGFJ01009375 MBW58516.1 GAKP01001359 JAC57593.1 GDHF01012481 JAI39833.1 GGFJ01009374 MBW58515.1 CM000157 EDW89600.1 GGFK01002294 MBW35615.1 CM000362 CM002911 EDX06440.1 KMY92659.1 AE013599 AY094698 AAF58856.1 AAM11051.1 GGFK01002296 MBW35617.1 CH902619 EDV36339.1 GFDL01009824 JAV25221.1 ADMH02002020 ETN59960.1 DS231987 EDS30533.1 CH480816 EDW47195.1 GBXI01003751 JAD10541.1 LBMM01000241 KMR03300.1 CH481236 EDW52800.1 JXJN01021173 JXJN01026673 GL732599 EFX72510.1
JAT88296.1 KQ458499 KPJ05858.1 KZ288189 PBC34666.1 KQ979608 KYN20474.1 KQ978324 KYM95109.1 KQ981965 KYN31839.1 ADTU01023744 KQ976691 KYM77913.1 GL765434 EFZ16278.1 KQ971351 EFA06585.2 KQ435794 KOX73850.1 GEZM01101845 GEZM01101844 JAV52257.1 UFQT01001052 SSX28651.1 NEVH01015304 PNF27174.1 GBYB01008700 JAG78467.1 NNAY01000791 OXU26490.1 GBRD01000242 JAG65579.1 KK107139 EZA57730.1 GFDF01003426 JAV10658.1 GFDF01003427 JAV10657.1 GFDF01003428 JAV10656.1 GFDF01003429 JAV10655.1 GBHO01025051 GDHC01004043 JAG18553.1 JAQ14586.1 CP012524 ALC41985.1 GECU01027658 JAS80048.1 CH916367 EDW01811.1 DQ440267 ABF18300.1 KB632333 ERL92671.1 CH963849 EDW74511.1 CH940648 EDW62233.2 APGK01035263 KB740923 ENN78236.1 JH431812 GFXV01001059 GFXV01002379 MBW12864.1 MBW14184.1 CM000071 EAL25277.1 CH479181 EDW32201.1 GAPW01004533 JAC09065.1 CH477959 EAT34790.1 JRES01000780 KNC28378.1 GGMS01017004 MBY86207.1 GECZ01024437 JAS45332.1 OUUW01000001 SPP73625.1 GAPW01004480 GAPW01004479 JAC09119.1 GFDG01000400 JAV18399.1 JXUM01046464 KQ561478 KXJ78464.1 QOIP01000003 RLU24944.1 GGFM01002964 MBW23715.1 GFTR01003147 JAW13279.1 CH933808 EDW08702.2 CH954177 EDV58166.1 GGFJ01009375 MBW58516.1 GAKP01001359 JAC57593.1 GDHF01012481 JAI39833.1 GGFJ01009374 MBW58515.1 CM000157 EDW89600.1 GGFK01002294 MBW35615.1 CM000362 CM002911 EDX06440.1 KMY92659.1 AE013599 AY094698 AAF58856.1 AAM11051.1 GGFK01002296 MBW35617.1 CH902619 EDV36339.1 GFDL01009824 JAV25221.1 ADMH02002020 ETN59960.1 DS231987 EDS30533.1 CH480816 EDW47195.1 GBXI01003751 JAD10541.1 LBMM01000241 KMR03300.1 CH481236 EDW52800.1 JXJN01021173 JXJN01026673 GL732599 EFX72510.1
Proteomes
UP000007151
UP000005204
UP000053268
UP000005203
UP000242457
UP000078492
+ More
UP000078542 UP000078541 UP000005205 UP000078540 UP000007266 UP000053105 UP000235965 UP000215335 UP000002358 UP000053097 UP000092553 UP000001070 UP000030742 UP000007798 UP000008792 UP000019118 UP000001819 UP000008744 UP000008820 UP000037069 UP000095301 UP000268350 UP000069940 UP000249989 UP000279307 UP000069272 UP000192221 UP000009192 UP000008711 UP000002282 UP000000304 UP000000803 UP000007801 UP000000673 UP000002320 UP000001292 UP000036403 UP000078200 UP000092443 UP000092460 UP000000305
UP000078542 UP000078541 UP000005205 UP000078540 UP000007266 UP000053105 UP000235965 UP000215335 UP000002358 UP000053097 UP000092553 UP000001070 UP000030742 UP000007798 UP000008792 UP000019118 UP000001819 UP000008744 UP000008820 UP000037069 UP000095301 UP000268350 UP000069940 UP000249989 UP000279307 UP000069272 UP000192221 UP000009192 UP000008711 UP000002282 UP000000304 UP000000803 UP000007801 UP000000673 UP000002320 UP000001292 UP000036403 UP000078200 UP000092443 UP000092460 UP000000305
Pfam
PF11938 DUF3456
ProteinModelPortal
A0A2H1VES3
A0A212ETX8
H9J180
A0A1E1WN67
A0A0N1I653
A0A088AH64
+ More
A0A2A3ETX6 A0A195E6J8 A0A151I932 A0A195EVC2 A0A158NR17 A0A195B0A9 E9ISN7 D6WRV5 A0A0M8ZYS0 A0A1Y1K0K3 A0A336MGW9 A0A2J7QF17 A0A0C9RKH5 A0A232F7L9 K7IYX5 A0A0K8TJB0 A0A026WNY7 A0A1L8DW48 A0A1L8DW36 A0A1L8DW46 A0A1L8DWL6 A0A0A9XIC9 A0A1S4FXN3 A0A0M4EHF4 A0A1B6HZD7 B4J5N5 Q1HR27 U4UR88 B4MQX2 B4LP39 N6UC79 T1J2X4 A0A2H8TFJ5 Q290T5 B4GB99 A0A023EJE0 Q16KG3 A0A0L0C7M6 A0A1I8MAC8 A0A2S2R826 A0A1B6F505 A0A3B0IZI8 A0A023ELJ2 A0A1L8EIG1 A0A182G7E1 A0A3L8DWU6 A0A2M3Z5D4 A0A224XYW7 A0A182F6A9 A0A1W4VDN5 B4KM38 B3N6J7 A0A2M4BZM3 A0A034WUZ1 A0A0K8VLW0 A0A2M4BZK8 B4NX39 A0A2M4A4D5 B4QI80 Q7JXF7 A0A2M4A491 B3MD24 A0A1Q3FCA7 W5JAU4 B0WLK3 B4HMB8 A0A0A1XHT4 A0A0J7L8K5 A0A1A9USS0 B4IMW9 A0A1A9YPB8 A0A1B0C6I0 E9H737
A0A2A3ETX6 A0A195E6J8 A0A151I932 A0A195EVC2 A0A158NR17 A0A195B0A9 E9ISN7 D6WRV5 A0A0M8ZYS0 A0A1Y1K0K3 A0A336MGW9 A0A2J7QF17 A0A0C9RKH5 A0A232F7L9 K7IYX5 A0A0K8TJB0 A0A026WNY7 A0A1L8DW48 A0A1L8DW36 A0A1L8DW46 A0A1L8DWL6 A0A0A9XIC9 A0A1S4FXN3 A0A0M4EHF4 A0A1B6HZD7 B4J5N5 Q1HR27 U4UR88 B4MQX2 B4LP39 N6UC79 T1J2X4 A0A2H8TFJ5 Q290T5 B4GB99 A0A023EJE0 Q16KG3 A0A0L0C7M6 A0A1I8MAC8 A0A2S2R826 A0A1B6F505 A0A3B0IZI8 A0A023ELJ2 A0A1L8EIG1 A0A182G7E1 A0A3L8DWU6 A0A2M3Z5D4 A0A224XYW7 A0A182F6A9 A0A1W4VDN5 B4KM38 B3N6J7 A0A2M4BZM3 A0A034WUZ1 A0A0K8VLW0 A0A2M4BZK8 B4NX39 A0A2M4A4D5 B4QI80 Q7JXF7 A0A2M4A491 B3MD24 A0A1Q3FCA7 W5JAU4 B0WLK3 B4HMB8 A0A0A1XHT4 A0A0J7L8K5 A0A1A9USS0 B4IMW9 A0A1A9YPB8 A0A1B0C6I0 E9H737
Ontologies
Topology
Subcellular location
Endoplasmic reticulum
SignalP
Position: 1 - 19,
Likelihood: 0.996869
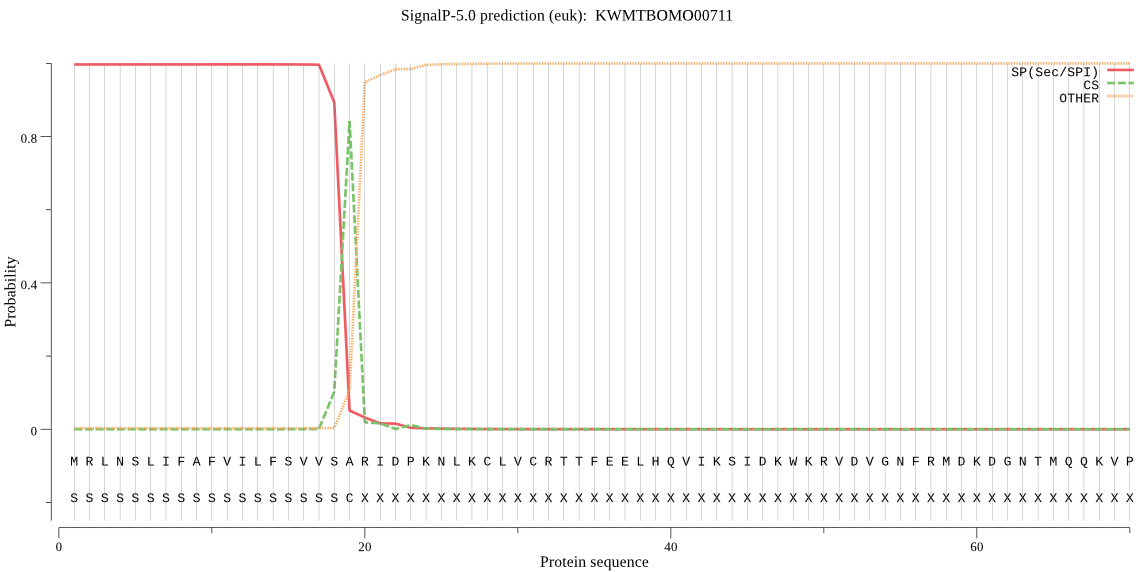
Length:
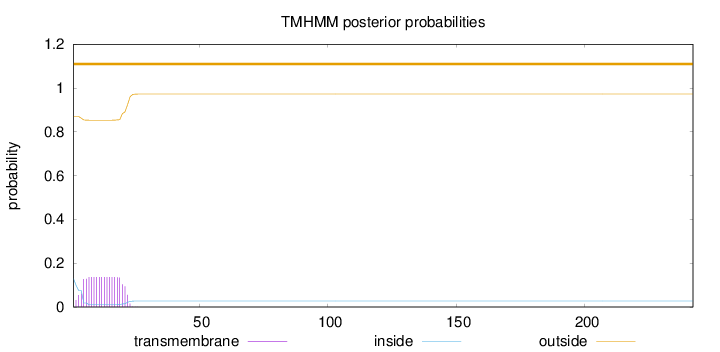
242
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.44256
Exp number, first 60 AAs:
2.44186
Total prob of N-in:
0.12993
outside
1 - 242
Population Genetic Test Statistics
Pi
301.419763
Theta
172.289105
Tajima's D
2.254836
CLR
0.229334
CSRT
0.91890405479726
Interpretation
Uncertain
