Gene
KWMTBOMO00710
Pre Gene Modal
BGIBMGA003229
Annotation
3-hydroxy-3-methylglutaryl-CoA_reductase_precursor_[Bombyx_mori]
Full name
3-hydroxy-3-methylglutaryl coenzyme A reductase
+ More
3-hydroxy-3-methylglutaryl-coenzyme A reductase
3-hydroxy-3-methylglutaryl-coenzyme A reductase
Location in the cell
PlasmaMembrane Reliability : 2.624
Sequence
CDS
ATGAAAGTGTGGGGAGCCCACGGGGAATTCTGCGCCAGACATCAATGGGAGGTCATCGTAGCCACCCTGGCTTTACTAGCCTGTGCAGCTAGTGTCGAGCGACATGGCCCAGGGAACAGATCTGAACACTGTGCTGGTTGGGCCCGAGCGTGTCCCGGTCTTGAAGCGGAGTATCAAGCAGCCGACGCGGTTATCATGACTTTTGTCCGCTGTGCCGCCCTGCTATATGCATACTACCAAGTTCTCAACCTTCACAAAATAGCCTCGAAGTACCTCCTCATCATAGCCGGGGTGTTCTCAACGTTTGCCAGTTTTATATTCACGTCGGCACTCGCTAGCCTGTTCTGGAGCGAATTGGCTAGTATAAAGGATGCGCCGTTTCTCTTTCTGTTGGTGGCTGATGTCGCCAGAGGTGCTAGGATGGCGAAAGCGGGGTGGTCGGCCGGTGAAGACCAAGGGAAGAGGGTTGGGAGGGCCTTGTCGTTGTTGGGACCAACTGCAACTCTAGACACACTGCTCGCCATATTATTGGTCGGCGTCGGAGCTTTATCCGGTGTTCCCCGTCTCGAGCATATGTGTACTTTTGCTTGCCTAGCCTTGTTGGTTGACTACATTGTGTTCATAACTTTCTACCCCGCTTGTTTGTCATTGGTAGCAGATTTCGCAACGAACAGAAAAGAAATAGCCCACGACAGTCCATTCTCGGAGGAGGACTTGAAACCCAACCCCGTGGTGCAGAGAGTCAAAATGATTATGGCCGCCGGCTTATTGTGCGTTCATTTGACGAGCAGGTGGCCTTGGTCTGCTAATCACGGCATAATTGAGGGACCAATTGATGCATCCATACCTGTTCCCCACGACAATATACTCCTCCATTCATACGTGAAGTGGTTCTCTGTGAGCGCAGACTACATCGTTATAGCGACGCTGCTCTGCGCTTTGATTATAAAATTCATTTTCTTTGAAGAGCAACGTAACTGGGTTTATGATATGGACGACATGACTGTAAAGGAAGTGATAAATGACACCGATTTCAGCCGTAAACCCAAGTTTTCTGTCGGTGACGATTCCAACTCTGAAGTATCCACACAGACCGACGAGGCTGGGAATGTAGAGGACATGGAGTGGCCCACTTTGTCCCCAAGCTCCTCGGCCTCTAAATTGAACGCCAAAAAGAGGCCTATGGTCGAATGCTTGGAGTTATATCGATCTGGTGCTTGCACTTCTCTCAGCGACGAAGAAGTCATAATGCTAGTCGAGCAATCCCATATACCCATGCACAGATTGGAAGCTGTGCTGGAGGATCCCTTGCGCGGTGTGAGACTGCGCCGGCGAGTGATCGCGTCGAGATTTAATAACGAAACCGCAATAAAACAACTTCCCTACTTAAACTACGATTATAGTAAAGTCTTAAACGCGTGCTGTGAGAATGTGATTGGCTACATTGGAGTTCCCGTCGGATATGCCGGCCCGTTGGTGGTAGACGGGAAGCCTTACATGATCCCGATGGCTACGACCGAGGGAGCTTTGGTGGCGTCAACAAACCGCGGAGCCAAAGCTATAGGCAGTAGGGGAGTGACGAGTGTCGTCGAAGATGTCGGCATGACAAGAGCGCCGGCTGTCAAATTGCCAAACGTAGTGCGAGCCCACGAATGCCGACAATGGATCGATAATAAAGAGAATTACGCTTTGCTCAAAGAAGCGTTCGATTCGACTTCAAGATTCGCCCGGCTCCAAGAAATACACGTCGGAGTTGATGGAGCTACTCTCTATCTAAGGTTCAGAGCGACAACTGGTGACGCTATGGGAATGAACATGGTCTCGAAAGGCGCGGAAAACGCTTTAAAACTATTAAAAACCTTCTTCCGTGATATGGAAGTTATTAGTTTGTCAGGGAATTATTGTTCGGACAAAAAGGCAGCGGCGATAAACTGGATCAAAGGTAGAGGTAAAAGGGTTGTCTGCGAGACTGTCATATCCAGTGAAAATTTGCGCACGATCTTCAAGACTGACGCCAAAACTCTGTCCCGGTGCAACAAAATCAAGAATTTATCCGGTTCCGCATTAGCCGGGTCTATAGGCGGCAACAACGCGCACGCGGCCAACATGGTTACCGCTATTTTCATAGCTACCGGCCAAGATCCGGCTCAGAACGTCACTAGCAGCAACTGCTCCACAAATATGGAAGCTTACGGAGAAAACGGCGAAGATTTATACGTTACCTGCACCATGCCCTCCTTGGAAGTCGGCACAGTAGGTGGGGGGACAGTTTTGACGGGTCAGGGAGCGTGTCTAGAAATCTTAGGAGTCAAAGGAGCGGGTACGCGACCTGCCGAGAACTCCGCCAGGCTGGCGTCCCTAATATGCGCTACGGTTTTAGCTGGCGAGCTTAGTCTAATGGCGGCCTTGGTTAATTCGGATCTGGTGAAATCGCACATGCGTCACAACAGATCGACGCTTAACGTTCAAACTGCCAATGTCGAACCTTACACGGTGGCACTCAAAGTACCTCCATCATAA
Protein
MKVWGAHGEFCARHQWEVIVATLALLACAASVERHGPGNRSEHCAGWARACPGLEAEYQAADAVIMTFVRCAALLYAYYQVLNLHKIASKYLLIIAGVFSTFASFIFTSALASLFWSELASIKDAPFLFLLVADVARGARMAKAGWSAGEDQGKRVGRALSLLGPTATLDTLLAILLVGVGALSGVPRLEHMCTFACLALLVDYIVFITFYPACLSLVADFATNRKEIAHDSPFSEEDLKPNPVVQRVKMIMAAGLLCVHLTSRWPWSANHGIIEGPIDASIPVPHDNILLHSYVKWFSVSADYIVIATLLCALIIKFIFFEEQRNWVYDMDDMTVKEVINDTDFSRKPKFSVGDDSNSEVSTQTDEAGNVEDMEWPTLSPSSSASKLNAKKRPMVECLELYRSGACTSLSDEEVIMLVEQSHIPMHRLEAVLEDPLRGVRLRRRVIASRFNNETAIKQLPYLNYDYSKVLNACCENVIGYIGVPVGYAGPLVVDGKPYMIPMATTEGALVASTNRGAKAIGSRGVTSVVEDVGMTRAPAVKLPNVVRAHECRQWIDNKENYALLKEAFDSTSRFARLQEIHVGVDGATLYLRFRATTGDAMGMNMVSKGAENALKLLKTFFRDMEVISLSGNYCSDKKAAAINWIKGRGKRVVCETVISSENLRTIFKTDAKTLSRCNKIKNLSGSALAGSIGGNNAHAANMVTAIFIATGQDPAQNVTSSNCSTNMEAYGENGEDLYVTCTMPSLEVGTVGGGTVLTGQGACLEILGVKGAGTRPAENSARLASLICATVLAGELSLMAALVNSDLVKSHMRHNRSTLNVQTANVEPYTVALKVPPS
Summary
Description
Synthesis of mevalonate for the production of non-sterol isoprenoids, which are essential for growth differentiation.
Synthesis of mevalonate for the production of non-sterol isoprenoids, which are essential for growth differentiation. Provides spatial information during embryogenesis to guide migrating primordial germ cells (the pole cells) from the ectoderm to the mesoderm. Also required for association of the pole cells with the gonadal mesoderm.
Synthesis of mevalonate for the production of non-sterol isoprenoids, which are essential for growth differentiation. Provides spatial information during embryogenesis to guide migrating primordial germ cells (the pole cells) from the ectoderm to the mesoderm. Also required for association of the pole cells with the gonadal mesoderm.
Catalytic Activity
(R)-mevalonate + CoA + 2 NADP(+) = (3S)-hydroxy-3-methylglutaryl-CoA + 2 H(+) + 2 NADPH
Subunit
Homodimer. Interacts with INSIG1 (via its SSD); the interaction, accelerated by sterols, leads to the recruitment of HMGCR to AMFR/gp78 for its ubiquitination by the sterol-mediated ERAD pathway. Interacts with UBIAD1 (By similarity).
Similarity
Belongs to the HMG-CoA reductase family.
Keywords
Endoplasmic reticulum
Glycoprotein
Isoprene biosynthesis
Membrane
NADP
Oxidoreductase
Transmembrane
Transmembrane helix
Complete proteome
Reference proteome
Cholesterol biosynthesis
Cholesterol metabolism
Isopeptide bond
Lipid biosynthesis
Lipid metabolism
Phosphoprotein
Steroid biosynthesis
Steroid metabolism
Sterol biosynthesis
Sterol metabolism
Ubl conjugation
Feature
chain 3-hydroxy-3-methylglutaryl coenzyme A reductase
Uniprot
A5A799
H9J142
A0A2A4JNE1
A0A2W1BG38
O76819
F8QQS1
+ More
A0A2H1VEX4 Q1KN65 A0A0L7LDS5 A0A212ERM5 A0A0N1PFF1 U3L2Y7 B4K5A6 E1JIU8 B4HFX8 P14773 A0A1W4UJE0 B4PNX0 A9UN11 B3P8E9 B3LY57 A0A0Q9XBM3 A0A0P8Y1C8 A0A0M4EWC0 B4NG69 A0A1L8DYC9 A0A1L8DYA1 A0A0L0CBV1 W8BBG5 A0A3B0JLC7 A0A0A1X7T9 B4M0G1 A0A1A9ZMA8 B4JT90 A0A034VAL2 A0A0K8UZR1 Q299D2 B4G517 Q16W73 A0A1Q3FAJ7 A0A0K8UCR3 B0WAH3 A0A1I8MX88 X4R2H0 A0A0A1XEH4 A0A1A9XKU1 A0A1B0AVB5 A0A2M4AIB4 A0A172DZL8 A0A0L7RBM5 A0A139WIA9 I1VX01 A0A0G3FAB1 A0A0H4ISG3 A0A0F7LG51 Q7QKE7 N6UHD7 T1PKH0 U5ZZ40 E0VQY1 Q9N6G8 A0A1W4X6G1 A0A182XVC9 A0A067R6C0 A4KXN8 A0A154PLS1 A0A2J7R3S8 D6WSF8 A0A0B4ZSX0 A0A182HS22 A0A336MWC9 A0A2R7VQZ9 A0A024BSN1 K0J2Y0 Q9XY99 A0A224XJR7 Q95WT1 A0A069DXA3 A0A1J1J8B8 Q9NDD2 N6UKH0 P54960 A0A1A9X2D3 A0A2C9JZ80 A0A1L8DYL0 A0A1L8HRD9 A0A1U8CMG6 A0A1U7Q6F6 P09610 A0A1Y1LJY8 A0A140CT20 A0A182FPY6 A0A2M4BDS5 A0A2M4BDW4 A0A2M4BDW5 A0A0H4IVW3 F6V051 A0A091KC81
A0A2H1VEX4 Q1KN65 A0A0L7LDS5 A0A212ERM5 A0A0N1PFF1 U3L2Y7 B4K5A6 E1JIU8 B4HFX8 P14773 A0A1W4UJE0 B4PNX0 A9UN11 B3P8E9 B3LY57 A0A0Q9XBM3 A0A0P8Y1C8 A0A0M4EWC0 B4NG69 A0A1L8DYC9 A0A1L8DYA1 A0A0L0CBV1 W8BBG5 A0A3B0JLC7 A0A0A1X7T9 B4M0G1 A0A1A9ZMA8 B4JT90 A0A034VAL2 A0A0K8UZR1 Q299D2 B4G517 Q16W73 A0A1Q3FAJ7 A0A0K8UCR3 B0WAH3 A0A1I8MX88 X4R2H0 A0A0A1XEH4 A0A1A9XKU1 A0A1B0AVB5 A0A2M4AIB4 A0A172DZL8 A0A0L7RBM5 A0A139WIA9 I1VX01 A0A0G3FAB1 A0A0H4ISG3 A0A0F7LG51 Q7QKE7 N6UHD7 T1PKH0 U5ZZ40 E0VQY1 Q9N6G8 A0A1W4X6G1 A0A182XVC9 A0A067R6C0 A4KXN8 A0A154PLS1 A0A2J7R3S8 D6WSF8 A0A0B4ZSX0 A0A182HS22 A0A336MWC9 A0A2R7VQZ9 A0A024BSN1 K0J2Y0 Q9XY99 A0A224XJR7 Q95WT1 A0A069DXA3 A0A1J1J8B8 Q9NDD2 N6UKH0 P54960 A0A1A9X2D3 A0A2C9JZ80 A0A1L8DYL0 A0A1L8HRD9 A0A1U8CMG6 A0A1U7Q6F6 P09610 A0A1Y1LJY8 A0A140CT20 A0A182FPY6 A0A2M4BDS5 A0A2M4BDW4 A0A2M4BDW5 A0A0H4IVW3 F6V051 A0A091KC81
EC Number
1.1.1.34
Pubmed
17628279
19121390
28756777
10971716
18839418
26227816
+ More
22118469 26354079 17994087 18057021 3136321 10731132 12537572 9853754 17550304 26108605 24495485 25830018 25348373 15632085 17510324 25315136 18362917 19820115 26899871 25983273 12364791 23537049 24167290 20566863 12581875 25244985 24845553 17296500 22984844 10065157 12046626 16640731 26334808 16933277 8477698 15562597 27762356 3841506 8473286 8288583 28004739 26850644 17495919
22118469 26354079 17994087 18057021 3136321 10731132 12537572 9853754 17550304 26108605 24495485 25830018 25348373 15632085 17510324 25315136 18362917 19820115 26899871 25983273 12364791 23537049 24167290 20566863 12581875 25244985 24845553 17296500 22984844 10065157 12046626 16640731 26334808 16933277 8477698 15562597 27762356 3841506 8473286 8288583 28004739 26850644 17495919
EMBL
AB274990
BAF62108.1
BABH01031435
BABH01031436
NWSH01000986
PCG73238.1
+ More
KZ150260 PZC71750.1 AJ009675 GU584103 ADM13643.1 ODYU01002169 SOQ39321.1 DQ465407 ABE98255.1 JTDY01001521 KOB73663.1 AGBW02013007 OWR44104.1 KQ460940 KPJ10638.1 JQ403110 AFV40971.1 CH933806 EDW16132.1 KRG01951.1 BT150309 AGW24091.1 CH480815 EDW43371.1 M21329 AE014297 CM000160 EDW98180.2 BT031175 ABY20416.1 CH954182 EDV53973.1 CH902617 EDV43961.1 KPU80562.1 KRG01950.1 KPU80561.1 CP012526 ALC47805.1 CH964251 EDW83286.1 GFDF01002636 JAV11448.1 GFDF01002637 JAV11447.1 JRES01000634 KNC29741.1 GAMC01010533 JAB96022.1 OUUW01000007 SPP83035.1 GBXI01006948 JAD07344.1 CH940650 EDW68340.1 KRF83787.1 CH916373 EDV94980.1 GAKP01020147 JAC38805.1 GDHF01020218 GDHF01008114 JAI32096.1 JAI44200.1 CM000070 EAL27771.1 CH479179 EDW24683.1 CH477573 EAT38837.1 GFDL01010517 JAV24528.1 GDHF01027956 GDHF01000671 JAI24358.1 JAI51643.1 DS231872 EDS41396.1 KF149986 AHU87084.1 GBXI01004942 JAD09350.1 JXJN01004170 GGFK01007200 MBW40521.1 JX096638 AGF87101.1 KQ414617 KOC68238.1 KQ971342 KYB27676.1 JQ413983 AFI55099.1 KP420145 AKJ80165.1 KP689337 AKO63319.1 KP317811 AKH60758.1 AAAB01008799 EAA03787.5 APGK01020365 KB740193 KB631602 ENN81135.1 ERL84534.1 KA648580 AFP63209.1 KF444677 AHA11094.1 DS235442 EEB15787.1 AH009515 AF159136 AAF80373.1 AAF80374.1 KK852921 KDR13783.1 EF134407 ABO37160.1 KQ434977 KZC12825.1 NEVH01007818 PNF35486.1 KQ971352 EFA06636.1 KM085007 AJD79560.1 APCN01000233 APCN01000234 UFQT01003361 SSX34904.1 KK854005 PTY09080.1 KJ188023 AHZ20731.1 AB733009 BAM45097.1 AF071750 AAD20975.2 GFTR01008165 JAW08261.1 AF304440 AAL09351.1 GBGD01000448 JAC88441.1 CVRI01000073 CRL07726.1 AF162705 AAF80475.2 ENN81136.1 X70034 GFDF01002729 JAV11355.1 CM004467 OCT98653.1 M12705 GEZM01057567 GEZM01057566 JAV72255.1 KU522123 AMH87089.1 GGFJ01002054 MBW51195.1 GGFJ01002093 MBW51234.1 GGFJ01002094 MBW51235.1 KP689336 AKO63318.1 KK737895 KFP37702.1
KZ150260 PZC71750.1 AJ009675 GU584103 ADM13643.1 ODYU01002169 SOQ39321.1 DQ465407 ABE98255.1 JTDY01001521 KOB73663.1 AGBW02013007 OWR44104.1 KQ460940 KPJ10638.1 JQ403110 AFV40971.1 CH933806 EDW16132.1 KRG01951.1 BT150309 AGW24091.1 CH480815 EDW43371.1 M21329 AE014297 CM000160 EDW98180.2 BT031175 ABY20416.1 CH954182 EDV53973.1 CH902617 EDV43961.1 KPU80562.1 KRG01950.1 KPU80561.1 CP012526 ALC47805.1 CH964251 EDW83286.1 GFDF01002636 JAV11448.1 GFDF01002637 JAV11447.1 JRES01000634 KNC29741.1 GAMC01010533 JAB96022.1 OUUW01000007 SPP83035.1 GBXI01006948 JAD07344.1 CH940650 EDW68340.1 KRF83787.1 CH916373 EDV94980.1 GAKP01020147 JAC38805.1 GDHF01020218 GDHF01008114 JAI32096.1 JAI44200.1 CM000070 EAL27771.1 CH479179 EDW24683.1 CH477573 EAT38837.1 GFDL01010517 JAV24528.1 GDHF01027956 GDHF01000671 JAI24358.1 JAI51643.1 DS231872 EDS41396.1 KF149986 AHU87084.1 GBXI01004942 JAD09350.1 JXJN01004170 GGFK01007200 MBW40521.1 JX096638 AGF87101.1 KQ414617 KOC68238.1 KQ971342 KYB27676.1 JQ413983 AFI55099.1 KP420145 AKJ80165.1 KP689337 AKO63319.1 KP317811 AKH60758.1 AAAB01008799 EAA03787.5 APGK01020365 KB740193 KB631602 ENN81135.1 ERL84534.1 KA648580 AFP63209.1 KF444677 AHA11094.1 DS235442 EEB15787.1 AH009515 AF159136 AAF80373.1 AAF80374.1 KK852921 KDR13783.1 EF134407 ABO37160.1 KQ434977 KZC12825.1 NEVH01007818 PNF35486.1 KQ971352 EFA06636.1 KM085007 AJD79560.1 APCN01000233 APCN01000234 UFQT01003361 SSX34904.1 KK854005 PTY09080.1 KJ188023 AHZ20731.1 AB733009 BAM45097.1 AF071750 AAD20975.2 GFTR01008165 JAW08261.1 AF304440 AAL09351.1 GBGD01000448 JAC88441.1 CVRI01000073 CRL07726.1 AF162705 AAF80475.2 ENN81136.1 X70034 GFDF01002729 JAV11355.1 CM004467 OCT98653.1 M12705 GEZM01057567 GEZM01057566 JAV72255.1 KU522123 AMH87089.1 GGFJ01002054 MBW51195.1 GGFJ01002093 MBW51234.1 GGFJ01002094 MBW51235.1 KP689336 AKO63318.1 KK737895 KFP37702.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000009192
+ More
UP000001292 UP000000803 UP000192221 UP000002282 UP000008711 UP000007801 UP000092553 UP000007798 UP000037069 UP000268350 UP000008792 UP000092445 UP000001070 UP000001819 UP000008744 UP000008820 UP000002320 UP000095301 UP000092443 UP000092460 UP000053825 UP000007266 UP000007062 UP000019118 UP000030742 UP000009046 UP000192223 UP000076408 UP000027135 UP000076502 UP000235965 UP000075840 UP000183832 UP000091820 UP000076420 UP000186698 UP000189706 UP000069272 UP000002280
UP000001292 UP000000803 UP000192221 UP000002282 UP000008711 UP000007801 UP000092553 UP000007798 UP000037069 UP000268350 UP000008792 UP000092445 UP000001070 UP000001819 UP000008744 UP000008820 UP000002320 UP000095301 UP000092443 UP000092460 UP000053825 UP000007266 UP000007062 UP000019118 UP000030742 UP000009046 UP000192223 UP000076408 UP000027135 UP000076502 UP000235965 UP000075840 UP000183832 UP000091820 UP000076420 UP000186698 UP000189706 UP000069272 UP000002280
Interpro
Gene 3D
ProteinModelPortal
A5A799
H9J142
A0A2A4JNE1
A0A2W1BG38
O76819
F8QQS1
+ More
A0A2H1VEX4 Q1KN65 A0A0L7LDS5 A0A212ERM5 A0A0N1PFF1 U3L2Y7 B4K5A6 E1JIU8 B4HFX8 P14773 A0A1W4UJE0 B4PNX0 A9UN11 B3P8E9 B3LY57 A0A0Q9XBM3 A0A0P8Y1C8 A0A0M4EWC0 B4NG69 A0A1L8DYC9 A0A1L8DYA1 A0A0L0CBV1 W8BBG5 A0A3B0JLC7 A0A0A1X7T9 B4M0G1 A0A1A9ZMA8 B4JT90 A0A034VAL2 A0A0K8UZR1 Q299D2 B4G517 Q16W73 A0A1Q3FAJ7 A0A0K8UCR3 B0WAH3 A0A1I8MX88 X4R2H0 A0A0A1XEH4 A0A1A9XKU1 A0A1B0AVB5 A0A2M4AIB4 A0A172DZL8 A0A0L7RBM5 A0A139WIA9 I1VX01 A0A0G3FAB1 A0A0H4ISG3 A0A0F7LG51 Q7QKE7 N6UHD7 T1PKH0 U5ZZ40 E0VQY1 Q9N6G8 A0A1W4X6G1 A0A182XVC9 A0A067R6C0 A4KXN8 A0A154PLS1 A0A2J7R3S8 D6WSF8 A0A0B4ZSX0 A0A182HS22 A0A336MWC9 A0A2R7VQZ9 A0A024BSN1 K0J2Y0 Q9XY99 A0A224XJR7 Q95WT1 A0A069DXA3 A0A1J1J8B8 Q9NDD2 N6UKH0 P54960 A0A1A9X2D3 A0A2C9JZ80 A0A1L8DYL0 A0A1L8HRD9 A0A1U8CMG6 A0A1U7Q6F6 P09610 A0A1Y1LJY8 A0A140CT20 A0A182FPY6 A0A2M4BDS5 A0A2M4BDW4 A0A2M4BDW5 A0A0H4IVW3 F6V051 A0A091KC81
A0A2H1VEX4 Q1KN65 A0A0L7LDS5 A0A212ERM5 A0A0N1PFF1 U3L2Y7 B4K5A6 E1JIU8 B4HFX8 P14773 A0A1W4UJE0 B4PNX0 A9UN11 B3P8E9 B3LY57 A0A0Q9XBM3 A0A0P8Y1C8 A0A0M4EWC0 B4NG69 A0A1L8DYC9 A0A1L8DYA1 A0A0L0CBV1 W8BBG5 A0A3B0JLC7 A0A0A1X7T9 B4M0G1 A0A1A9ZMA8 B4JT90 A0A034VAL2 A0A0K8UZR1 Q299D2 B4G517 Q16W73 A0A1Q3FAJ7 A0A0K8UCR3 B0WAH3 A0A1I8MX88 X4R2H0 A0A0A1XEH4 A0A1A9XKU1 A0A1B0AVB5 A0A2M4AIB4 A0A172DZL8 A0A0L7RBM5 A0A139WIA9 I1VX01 A0A0G3FAB1 A0A0H4ISG3 A0A0F7LG51 Q7QKE7 N6UHD7 T1PKH0 U5ZZ40 E0VQY1 Q9N6G8 A0A1W4X6G1 A0A182XVC9 A0A067R6C0 A4KXN8 A0A154PLS1 A0A2J7R3S8 D6WSF8 A0A0B4ZSX0 A0A182HS22 A0A336MWC9 A0A2R7VQZ9 A0A024BSN1 K0J2Y0 Q9XY99 A0A224XJR7 Q95WT1 A0A069DXA3 A0A1J1J8B8 Q9NDD2 N6UKH0 P54960 A0A1A9X2D3 A0A2C9JZ80 A0A1L8DYL0 A0A1L8HRD9 A0A1U8CMG6 A0A1U7Q6F6 P09610 A0A1Y1LJY8 A0A140CT20 A0A182FPY6 A0A2M4BDS5 A0A2M4BDW4 A0A2M4BDW5 A0A0H4IVW3 F6V051 A0A091KC81
PDB
1HWL
E-value=2.12282e-124,
Score=1144
Ontologies
PATHWAY
GO
GO:0050661
GO:0004420
GO:0016021
GO:0005789
GO:0015936
GO:0008299
GO:0018990
GO:0035050
GO:0007626
GO:0035232
GO:0007280
GO:0046579
GO:0008406
GO:0005778
GO:0008354
GO:0042282
GO:0016126
GO:1900222
GO:0070402
GO:0042177
GO:0008542
GO:0006695
GO:0043407
GO:0051262
GO:0050709
GO:0016616
GO:0050662
GO:0006559
GO:0004887
GO:0055114
GO:0005515
PANTHER
Topology
Subcellular location
Endoplasmic reticulum membrane
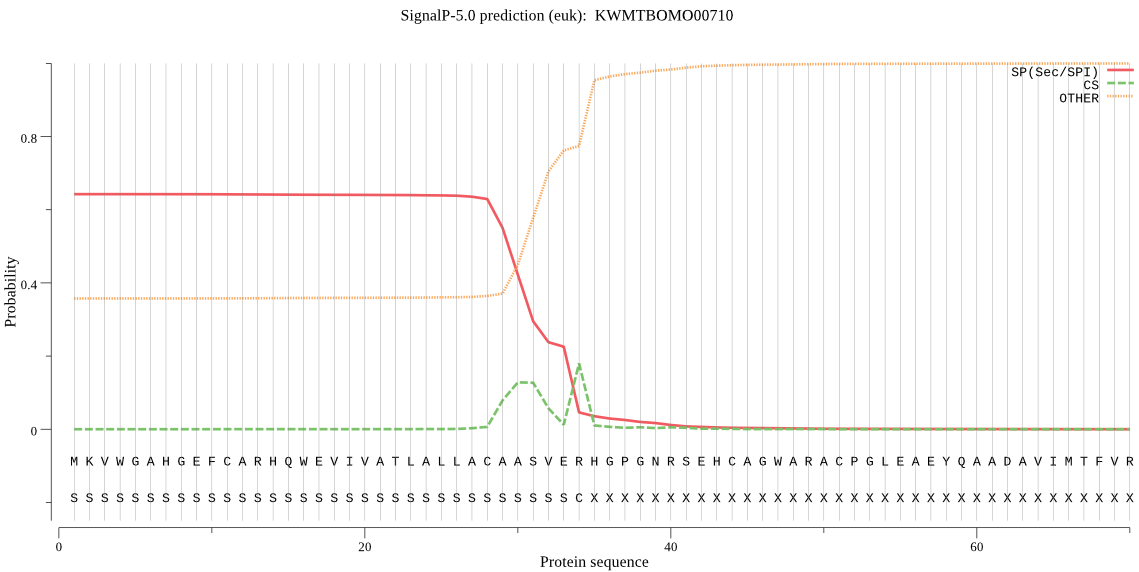
SignalP
Position: 1 - 34,
Likelihood: 0.642868
Length:
839
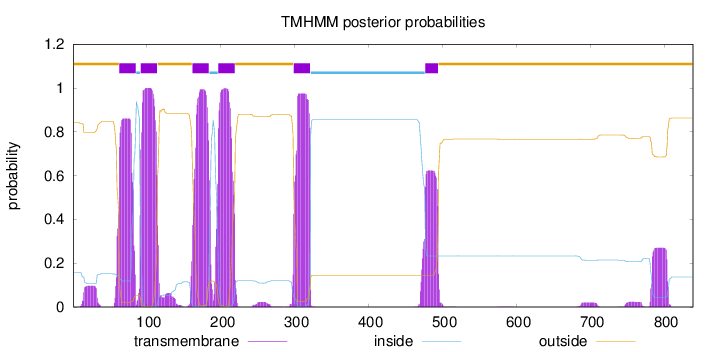
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
128.7238
Exp number, first 60 AAs:
2.47588
Total prob of N-in:
0.15807
outside
1 - 62
TMhelix
63 - 85
inside
86 - 91
TMhelix
92 - 114
outside
115 - 161
TMhelix
162 - 184
inside
185 - 196
TMhelix
197 - 219
outside
220 - 298
TMhelix
299 - 321
inside
322 - 476
TMhelix
477 - 494
outside
495 - 839
Population Genetic Test Statistics
Pi
289.119859
Theta
182.562176
Tajima's D
1.505151
CLR
0.049497
CSRT
0.789310534473276
Interpretation
Uncertain
