Gene
KWMTBOMO00709
Pre Gene Modal
BGIBMGA003268
Annotation
PREDICTED:_chaoptin_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.913
Sequence
CDS
ATGGTCTACACAATTGTATTTTTTGTATTAGCAACACTGCCACTTTTGAATTCAGAATACATTCCACCCGGTCCTGCATATTCATGCCCGAAGGAGACAACATTATTGTATCCCTGTATTTGTGAGAAAGGAACAGATTCAGGGCTGTATATACGTTGTGAAAATGCAGGTTTGGCTCTTATCAGTGTGGGAATTGGCAACATAGCTGGTATGGGGTTGCCAATAGAGAGACTGTTGCTGAAGGAATGCAAATTTACAAACCTCTTTGGTTCATTACTCCATAGATCGACTGTACGTATCTTGGAGATAGTGGACACACCTATTACGAGCATTGAAGAGTACGCTTTTGCTGGTGTAAACAGAACATTGCAGGAGATATACTTGACAAATACCAAGCTTGTCGATTTTCCTAAGGAGGCTTTTAAGATCTTGGGCAACCTTTCAGTATTGTCTATTGATGGACACCAAATGACTAAATTGAGTAAGGACATGTTCGCAGGCAGTGAAGCTGTGGGAAAACTGGAGAGATTACACCTTGTTAACGGCTTAATATCTGATATACCCATAGAAGCATTTCAGAACCTGCGTAAGTTAAAAAAGTTAGATCTGCACGGAAACAGGCTTGTGTCCTTGAAGAAGAATCAGTTCAAGGGACTCCGGGACACCGATGTCTTGGATCTGAGTCACAACAATATCACGAGGCTGGACGGCACTCACATCGGGGACCTGACCAAGTTGGGCTGGTGCAACGTCAGCCACAACGACATCAAGGAAATACCCCGGGGGATGTTCGCGAGGAACAATCTACTGAAAGTCCTGCATTTGGATAATAATAAATTGAAGAAATTGGACACGAATAGTTTCCGAGGAATGAGGTTTCTGAGGCGTTTATTCCTTCAAGACAATCAGATCTCGGATATAGGCAGGGGGACCTTCTCGGCCGTGACCAGAATAGGCACCGTGGACCTGGCGAGGAATAAAATCAAGAAAGTCGACTATCAAATGTTTCAATCGGTCAAATTTGCTGACACGATAAACTTGGCGGAGAACGAGATAACCGTGATCGAAGCCCAGGCTTTCACGGACCTGTATCTGGCGACGGTGAACGTCTCCCACAACCAGCTCTCCAAGATCGAACCGAAGGCCTTCCAGAACTGCAACAACCTGACCGTGCTGGACTTGCGCTACAACAACCTGACGGTCATCGATAGGAGCGCCTTCGACGAGAACACATACGCGTATGAATTGAATGTCGCCCACAACCACATCACGGATTTCAGTCAGATCCCCATACAGAACATGACGGGCATAAGGATCTTGAATGCGACCTTCAACCAGATCACCGGGATACCGAAAGCGGCGTTTCCCAAACTGTACGAGCTCCACACCGTCGATCTGTCACACAACAACATCAGCGACATCTTCAATGCGGTTTTCCAGAACCTGTTCTCCTTGAGATTCGTGAATCTGAGCCACAACAGCTTGGAGCGCATAAAACCCGCCATGTTCGGCACCCTACCCACCGTCTTGGAGCTGGATCTGTCGCACAACCTGCTGAGTGACGTGTCGAGGGGCGGGCTCGCCAAGCTGGCCAGCTGCCGCCTCCTGGACATGAGCCACAATCAACTGGAGAAGATATTCCAGATACCCATCAGTCTGGGCGAATTGAACGTAGCGCATAACAATCTGAGAGAGATAAAATCGAACACGTGGCCTTCGATGAACGCTCTGCTGCGTCTCAACCTGTCGGCCAACATGCTCGGCGACATGCTCGCCAGCGACAGCTTCTCCAGTCTGCTGACCTTGCAGACTCTGGACTTATCATCGAACGGGCTCACCAGACCGCCGTGGGAGGCCCTGAGCAGTCTGACGAGCTTACAGTATTTATATATGCAGGATAACAATCTGACGTCACTAAGCAAGTCGTCATTTGGAAACCTGCCCACGCTATTCGCGCTGGACCTGTCGCGCAACGCCATAGCTTCTGTGGAGCGCCGCGCTTTCAATGGCCTGCAGCAGCTACTGGACCTCAGGCTGTCAGGGAATCAACTGGAGCACGTCCCCAACGAGGCTTTTAAGGGCCTGGTAGCGCTCAGGAAGCTCGACCTGTCTCACAACTTACTGGAGAAGATCGACAACAAAACGAACGGACTTCTGGATGACTGTCTGTCCTTGGAGACGGTCAACTTGAGTCACAACAACATCGGCTTCCTAACGAAGAAGATGTTCCCGGCGAGCCCCTGGATCCCGTACAGACTGTCGGAGATCGACCTGAGTCACAACTCGCTGGCGGTCGTCACGCACGACATAACGGTGGGCGCGGGGCGCGTGCGGAGGCTGAGTCTGGCCGGCAATTACATCAACGACATCAGGAACAATGTCCTGGGCAACCTGACCTCGCTGGAGACCTTGGACCTTTCGCGGAACAACCTCAAGGACTTAGTGTCTAAAACATCGGAAATGAAATTCTCTTTCCCCGGGAACATAACAGAGCTGTATCTGAATGACAACGAGCTGGAGGATCTCCCGATCAAGGAGCTGATGAAGGCTGACAAACTGACGGTGCTGGACGTCAGGGGCAACCGGTTAACCAGCTTCGACCCCCGCCTGGTCGCGAGGGTCCGAGACCGGGGCTTGCAGGTCTACTTTGAAGGCAACACTTTGTTATGCGACTGCTTCGTTCGGCCCCTGAAGCATTACTTAAACAAACTGTCCCAGACGTACTTGCGCGGCGACGCTAAGTACTCAAACGTGAGCTGCGCTGAGCCTCCGAGCTTGGAGAACGTGAGCATATTGGACGTGGACGAGGAGAGGCTGCTGTGCGCTGGCAACATTGAGACCGACGAGAAGATAAAAGAATATGGCAACACCGCGGACGAGTCATTCAATTTCATAACGGAACCAGACTTGGCCTTCAGGGACATACAGTATTCCCAGGAGGCGATCTACGTGCACTGGTACGTGAGGTCGGCGGGGGATGTAGCCGACACGCAGCTATACGTGCGGGGGCAGGGGGGGGACGTGCTGCACGCCGCGCGCGCCCCCTACACCGCGCGCTCCTACACCGTCCCCCTGGACGCGGGGGTGCGAGGGGCGTTGGGGGGAGGCGGCGCGCTGTGCGTGCAAGCCGTGACGTCAGCGGGCGCGCCCCGACGCTGGTACCCCTCGCAGTGCCAACCAATAAACGGGCGTCTCCACCTATGGCCCAAGACGCTAAACGTGGATAAGCGGAGACTGTCCCGGCCCAGGGGACGCGCCACTAGCAGTGTCCTGGGACTCGTCAGCGACTCTATCCTGATCACGCTGTGCATATTCCTGGGGGTCTGTCTGAAGATAACCGACGTTGAGCTATGA
Protein
MVYTIVFFVLATLPLLNSEYIPPGPAYSCPKETTLLYPCICEKGTDSGLYIRCENAGLALISVGIGNIAGMGLPIERLLLKECKFTNLFGSLLHRSTVRILEIVDTPITSIEEYAFAGVNRTLQEIYLTNTKLVDFPKEAFKILGNLSVLSIDGHQMTKLSKDMFAGSEAVGKLERLHLVNGLISDIPIEAFQNLRKLKKLDLHGNRLVSLKKNQFKGLRDTDVLDLSHNNITRLDGTHIGDLTKLGWCNVSHNDIKEIPRGMFARNNLLKVLHLDNNKLKKLDTNSFRGMRFLRRLFLQDNQISDIGRGTFSAVTRIGTVDLARNKIKKVDYQMFQSVKFADTINLAENEITVIEAQAFTDLYLATVNVSHNQLSKIEPKAFQNCNNLTVLDLRYNNLTVIDRSAFDENTYAYELNVAHNHITDFSQIPIQNMTGIRILNATFNQITGIPKAAFPKLYELHTVDLSHNNISDIFNAVFQNLFSLRFVNLSHNSLERIKPAMFGTLPTVLELDLSHNLLSDVSRGGLAKLASCRLLDMSHNQLEKIFQIPISLGELNVAHNNLREIKSNTWPSMNALLRLNLSANMLGDMLASDSFSSLLTLQTLDLSSNGLTRPPWEALSSLTSLQYLYMQDNNLTSLSKSSFGNLPTLFALDLSRNAIASVERRAFNGLQQLLDLRLSGNQLEHVPNEAFKGLVALRKLDLSHNLLEKIDNKTNGLLDDCLSLETVNLSHNNIGFLTKKMFPASPWIPYRLSEIDLSHNSLAVVTHDITVGAGRVRRLSLAGNYINDIRNNVLGNLTSLETLDLSRNNLKDLVSKTSEMKFSFPGNITELYLNDNELEDLPIKELMKADKLTVLDVRGNRLTSFDPRLVARVRDRGLQVYFEGNTLLCDCFVRPLKHYLNKLSQTYLRGDAKYSNVSCAEPPSLENVSILDVDEERLLCAGNIETDEKIKEYGNTADESFNFITEPDLAFRDIQYSQEAIYVHWYVRSAGDVADTQLYVRGQGGDVLHAARAPYTARSYTVPLDAGVRGALGGGGALCVQAVTSAGAPRRWYPSQCQPINGRLHLWPKTLNVDKRRLSRPRGRATSSVLGLVSDSILITLCIFLGVCLKITDVEL
Summary
Uniprot
A0A2H1WSK0
A0A194PTT8
A0A0N1I7S4
A0A182JPP1
A0A182RCS2
D6WRC5
+ More
Q17FN7 A0A182L1D1 A0A182W701 A0A1S4GY69 A0A182WTZ7 A0A182VPG8 A0A182HUL3 A0A182GFS2 A0A182F4X7 A0A067R7C0 A0A182PPL4 W5JBW8 A0A2A3EMV7 E9IIM4 E2A1E1 A0A182NUG8 A0A0J7L688 A0A084W849 A0A182JDU2 A0A182QYT6 A0A195DP96 A0A087ZSD7 A0A195EZI1 A0A3L8E4G3 A0A195BID6 A0A182YQY2 A0A2J7R0N6 A0A158P3E5 A0A195CD70 A0A1Y1L0K3 E2C3L2 A0A2M4CN73 A0A182MB47 B4LMH6 A0A154P9W1 B4J759 B4KSK7 A0A1J1INW7 A0A232FGH1 A0A0L7RCV0 B4P737 A0A1I8MXN6 A0A0A1XB12 A0A1W4VZ34 A0A0M4E5P9 B3NR63 A1Z9N6 A0A1A9WIB3 A0A1B0AM45 A0A1A9XPR1 A0A1A9ZPC2 A0A1A9V7P4 A0A1B0GC25 A0A0M9A817 B4MJM4 B0WCT3 B3MDZ0 A0A026W3Z2 A0A3B0JMW2 B4QFF4 A0A0J9RCL8 A0A182TL04 K7J6D6 A0A0L0CB48 A0A0R3NVA5 Q28X54 A0A0K8W2Z1 A0A224XH39 A0A1W4XAP3 T1PHM6 W8AUG0 A0A151WYU6 T1IE03 A0A310SIP9 A0A182SZ20 E0VA95 Q7Q417 A0A1I8QCQ5 B4H518 A0A0P6HIA9 B4HRA7 A0A164YBP7 A0A0P5GME1 A0A0P6CIU7 A0A0P5UC61 A0A0P5SQ96 F4W4K3 A0A2S2R5K6 A0A2H8TU71 A0A0P5VL85
Q17FN7 A0A182L1D1 A0A182W701 A0A1S4GY69 A0A182WTZ7 A0A182VPG8 A0A182HUL3 A0A182GFS2 A0A182F4X7 A0A067R7C0 A0A182PPL4 W5JBW8 A0A2A3EMV7 E9IIM4 E2A1E1 A0A182NUG8 A0A0J7L688 A0A084W849 A0A182JDU2 A0A182QYT6 A0A195DP96 A0A087ZSD7 A0A195EZI1 A0A3L8E4G3 A0A195BID6 A0A182YQY2 A0A2J7R0N6 A0A158P3E5 A0A195CD70 A0A1Y1L0K3 E2C3L2 A0A2M4CN73 A0A182MB47 B4LMH6 A0A154P9W1 B4J759 B4KSK7 A0A1J1INW7 A0A232FGH1 A0A0L7RCV0 B4P737 A0A1I8MXN6 A0A0A1XB12 A0A1W4VZ34 A0A0M4E5P9 B3NR63 A1Z9N6 A0A1A9WIB3 A0A1B0AM45 A0A1A9XPR1 A0A1A9ZPC2 A0A1A9V7P4 A0A1B0GC25 A0A0M9A817 B4MJM4 B0WCT3 B3MDZ0 A0A026W3Z2 A0A3B0JMW2 B4QFF4 A0A0J9RCL8 A0A182TL04 K7J6D6 A0A0L0CB48 A0A0R3NVA5 Q28X54 A0A0K8W2Z1 A0A224XH39 A0A1W4XAP3 T1PHM6 W8AUG0 A0A151WYU6 T1IE03 A0A310SIP9 A0A182SZ20 E0VA95 Q7Q417 A0A1I8QCQ5 B4H518 A0A0P6HIA9 B4HRA7 A0A164YBP7 A0A0P5GME1 A0A0P6CIU7 A0A0P5UC61 A0A0P5SQ96 F4W4K3 A0A2S2R5K6 A0A2H8TU71 A0A0P5VL85
Pubmed
26354079
18362917
19820115
17510324
20966253
12364791
+ More
26483478 24845553 20920257 23761445 21282665 20798317 24438588 30249741 25244985 21347285 28004739 17994087 28648823 17550304 25315136 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19171940 26109357 26109356 24508170 22936249 20075255 26108605 15632085 24495485 20566863 21719571
26483478 24845553 20920257 23761445 21282665 20798317 24438588 30249741 25244985 21347285 28004739 17994087 28648823 17550304 25315136 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19171940 26109357 26109356 24508170 22936249 20075255 26108605 15632085 24495485 20566863 21719571
EMBL
ODYU01010707
SOQ55987.1
KQ459593
KPI96732.1
KQ460940
KPJ10635.1
+ More
KQ971351 EFA07490.2 CH477269 EAT45387.2 AAAB01008964 APCN01002465 JXUM01060201 JXUM01060202 JXUM01060203 JXUM01060204 JXUM01060205 JXUM01060206 JXUM01060207 JXUM01060208 JXUM01060209 KQ562092 KXJ76711.1 KK852657 KDR19221.1 ADMH02001944 ETN60390.1 KZ288204 PBC33143.1 GL763494 EFZ19579.1 GL435760 EFN72767.1 LBMM01000515 KMQ98161.1 ATLV01021362 KE525317 KFB46393.1 AXCN02000760 KQ980713 KYN14309.1 KQ981905 KYN33536.1 QOIP01000001 RLU27119.1 KQ976467 KYM84132.1 NEVH01008227 PNF34397.1 ADTU01007718 KQ977935 KYM98660.1 GEZM01070752 JAV66301.1 GL452320 EFN77550.1 GGFL01002547 MBW66725.1 AXCM01012047 CH940648 EDW62071.2 KQ434853 KZC08623.1 CH916367 EDW02077.1 CH933808 EDW09512.2 CVRI01000054 CRL00177.1 NNAY01000230 OXU29844.1 KQ414615 KOC68629.1 CM000158 EDW91002.2 GBXI01006156 JAD08136.1 CP012524 ALC41654.1 CH954179 EDV56052.2 AE013599 EU814872 AAF58265.2 ACI23397.1 AHN56214.1 JXJN01000330 CCAG010018974 KQ435714 KOX79145.1 CH963846 EDW72313.2 DS231890 EDS43809.1 CH902619 EDV37535.2 KK107447 EZA50723.1 OUUW01000001 SPP74656.1 CM000362 EDX07064.1 CM002911 KMY93737.1 JRES01000644 KNC29643.1 CM000071 KRT03117.1 EAL26462.3 GDHF01006838 JAI45476.1 GFTR01008534 JAW07892.1 KA648271 AFP62900.1 GAMC01016643 JAB89912.1 KQ982649 KYQ53053.1 ACPB03019348 KQ764883 OAD54292.1 DS235005 EEB10301.1 EAA12341.2 CH479210 EDW32854.1 GDIQ01019218 JAN75519.1 CH480816 EDW47836.1 LRGB01000930 KZS15088.1 GDIQ01238327 JAK13398.1 GDIP01007816 JAM95899.1 GDIP01115178 JAL88536.1 GDIP01141922 JAL61792.1 GL887532 EGI70898.1 GGMS01016031 MBY85234.1 GFXV01005556 MBW17361.1 GDIP01098470 JAM05245.1
KQ971351 EFA07490.2 CH477269 EAT45387.2 AAAB01008964 APCN01002465 JXUM01060201 JXUM01060202 JXUM01060203 JXUM01060204 JXUM01060205 JXUM01060206 JXUM01060207 JXUM01060208 JXUM01060209 KQ562092 KXJ76711.1 KK852657 KDR19221.1 ADMH02001944 ETN60390.1 KZ288204 PBC33143.1 GL763494 EFZ19579.1 GL435760 EFN72767.1 LBMM01000515 KMQ98161.1 ATLV01021362 KE525317 KFB46393.1 AXCN02000760 KQ980713 KYN14309.1 KQ981905 KYN33536.1 QOIP01000001 RLU27119.1 KQ976467 KYM84132.1 NEVH01008227 PNF34397.1 ADTU01007718 KQ977935 KYM98660.1 GEZM01070752 JAV66301.1 GL452320 EFN77550.1 GGFL01002547 MBW66725.1 AXCM01012047 CH940648 EDW62071.2 KQ434853 KZC08623.1 CH916367 EDW02077.1 CH933808 EDW09512.2 CVRI01000054 CRL00177.1 NNAY01000230 OXU29844.1 KQ414615 KOC68629.1 CM000158 EDW91002.2 GBXI01006156 JAD08136.1 CP012524 ALC41654.1 CH954179 EDV56052.2 AE013599 EU814872 AAF58265.2 ACI23397.1 AHN56214.1 JXJN01000330 CCAG010018974 KQ435714 KOX79145.1 CH963846 EDW72313.2 DS231890 EDS43809.1 CH902619 EDV37535.2 KK107447 EZA50723.1 OUUW01000001 SPP74656.1 CM000362 EDX07064.1 CM002911 KMY93737.1 JRES01000644 KNC29643.1 CM000071 KRT03117.1 EAL26462.3 GDHF01006838 JAI45476.1 GFTR01008534 JAW07892.1 KA648271 AFP62900.1 GAMC01016643 JAB89912.1 KQ982649 KYQ53053.1 ACPB03019348 KQ764883 OAD54292.1 DS235005 EEB10301.1 EAA12341.2 CH479210 EDW32854.1 GDIQ01019218 JAN75519.1 CH480816 EDW47836.1 LRGB01000930 KZS15088.1 GDIQ01238327 JAK13398.1 GDIP01007816 JAM95899.1 GDIP01115178 JAL88536.1 GDIP01141922 JAL61792.1 GL887532 EGI70898.1 GGMS01016031 MBY85234.1 GFXV01005556 MBW17361.1 GDIP01098470 JAM05245.1
Proteomes
UP000053268
UP000053240
UP000075881
UP000075900
UP000007266
UP000008820
+ More
UP000075882 UP000075920 UP000076407 UP000075903 UP000075840 UP000069940 UP000249989 UP000069272 UP000027135 UP000075885 UP000000673 UP000242457 UP000000311 UP000075884 UP000036403 UP000030765 UP000075880 UP000075886 UP000078492 UP000005203 UP000078541 UP000279307 UP000078540 UP000076408 UP000235965 UP000005205 UP000078542 UP000008237 UP000075883 UP000008792 UP000076502 UP000001070 UP000009192 UP000183832 UP000215335 UP000053825 UP000002282 UP000095301 UP000192221 UP000092553 UP000008711 UP000000803 UP000091820 UP000092460 UP000092443 UP000092445 UP000078200 UP000092444 UP000053105 UP000007798 UP000002320 UP000007801 UP000053097 UP000268350 UP000000304 UP000075902 UP000002358 UP000037069 UP000001819 UP000192223 UP000075809 UP000015103 UP000075901 UP000009046 UP000007062 UP000095300 UP000008744 UP000001292 UP000076858 UP000007755
UP000075882 UP000075920 UP000076407 UP000075903 UP000075840 UP000069940 UP000249989 UP000069272 UP000027135 UP000075885 UP000000673 UP000242457 UP000000311 UP000075884 UP000036403 UP000030765 UP000075880 UP000075886 UP000078492 UP000005203 UP000078541 UP000279307 UP000078540 UP000076408 UP000235965 UP000005205 UP000078542 UP000008237 UP000075883 UP000008792 UP000076502 UP000001070 UP000009192 UP000183832 UP000215335 UP000053825 UP000002282 UP000095301 UP000192221 UP000092553 UP000008711 UP000000803 UP000091820 UP000092460 UP000092443 UP000092445 UP000078200 UP000092444 UP000053105 UP000007798 UP000002320 UP000007801 UP000053097 UP000268350 UP000000304 UP000075902 UP000002358 UP000037069 UP000001819 UP000192223 UP000075809 UP000015103 UP000075901 UP000009046 UP000007062 UP000095300 UP000008744 UP000001292 UP000076858 UP000007755
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1WSK0
A0A194PTT8
A0A0N1I7S4
A0A182JPP1
A0A182RCS2
D6WRC5
+ More
Q17FN7 A0A182L1D1 A0A182W701 A0A1S4GY69 A0A182WTZ7 A0A182VPG8 A0A182HUL3 A0A182GFS2 A0A182F4X7 A0A067R7C0 A0A182PPL4 W5JBW8 A0A2A3EMV7 E9IIM4 E2A1E1 A0A182NUG8 A0A0J7L688 A0A084W849 A0A182JDU2 A0A182QYT6 A0A195DP96 A0A087ZSD7 A0A195EZI1 A0A3L8E4G3 A0A195BID6 A0A182YQY2 A0A2J7R0N6 A0A158P3E5 A0A195CD70 A0A1Y1L0K3 E2C3L2 A0A2M4CN73 A0A182MB47 B4LMH6 A0A154P9W1 B4J759 B4KSK7 A0A1J1INW7 A0A232FGH1 A0A0L7RCV0 B4P737 A0A1I8MXN6 A0A0A1XB12 A0A1W4VZ34 A0A0M4E5P9 B3NR63 A1Z9N6 A0A1A9WIB3 A0A1B0AM45 A0A1A9XPR1 A0A1A9ZPC2 A0A1A9V7P4 A0A1B0GC25 A0A0M9A817 B4MJM4 B0WCT3 B3MDZ0 A0A026W3Z2 A0A3B0JMW2 B4QFF4 A0A0J9RCL8 A0A182TL04 K7J6D6 A0A0L0CB48 A0A0R3NVA5 Q28X54 A0A0K8W2Z1 A0A224XH39 A0A1W4XAP3 T1PHM6 W8AUG0 A0A151WYU6 T1IE03 A0A310SIP9 A0A182SZ20 E0VA95 Q7Q417 A0A1I8QCQ5 B4H518 A0A0P6HIA9 B4HRA7 A0A164YBP7 A0A0P5GME1 A0A0P6CIU7 A0A0P5UC61 A0A0P5SQ96 F4W4K3 A0A2S2R5K6 A0A2H8TU71 A0A0P5VL85
Q17FN7 A0A182L1D1 A0A182W701 A0A1S4GY69 A0A182WTZ7 A0A182VPG8 A0A182HUL3 A0A182GFS2 A0A182F4X7 A0A067R7C0 A0A182PPL4 W5JBW8 A0A2A3EMV7 E9IIM4 E2A1E1 A0A182NUG8 A0A0J7L688 A0A084W849 A0A182JDU2 A0A182QYT6 A0A195DP96 A0A087ZSD7 A0A195EZI1 A0A3L8E4G3 A0A195BID6 A0A182YQY2 A0A2J7R0N6 A0A158P3E5 A0A195CD70 A0A1Y1L0K3 E2C3L2 A0A2M4CN73 A0A182MB47 B4LMH6 A0A154P9W1 B4J759 B4KSK7 A0A1J1INW7 A0A232FGH1 A0A0L7RCV0 B4P737 A0A1I8MXN6 A0A0A1XB12 A0A1W4VZ34 A0A0M4E5P9 B3NR63 A1Z9N6 A0A1A9WIB3 A0A1B0AM45 A0A1A9XPR1 A0A1A9ZPC2 A0A1A9V7P4 A0A1B0GC25 A0A0M9A817 B4MJM4 B0WCT3 B3MDZ0 A0A026W3Z2 A0A3B0JMW2 B4QFF4 A0A0J9RCL8 A0A182TL04 K7J6D6 A0A0L0CB48 A0A0R3NVA5 Q28X54 A0A0K8W2Z1 A0A224XH39 A0A1W4XAP3 T1PHM6 W8AUG0 A0A151WYU6 T1IE03 A0A310SIP9 A0A182SZ20 E0VA95 Q7Q417 A0A1I8QCQ5 B4H518 A0A0P6HIA9 B4HRA7 A0A164YBP7 A0A0P5GME1 A0A0P6CIU7 A0A0P5UC61 A0A0P5SQ96 F4W4K3 A0A2S2R5K6 A0A2H8TU71 A0A0P5VL85
PDB
6GFF
E-value=1.59148e-24,
Score=284
Ontologies
GO
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.993133
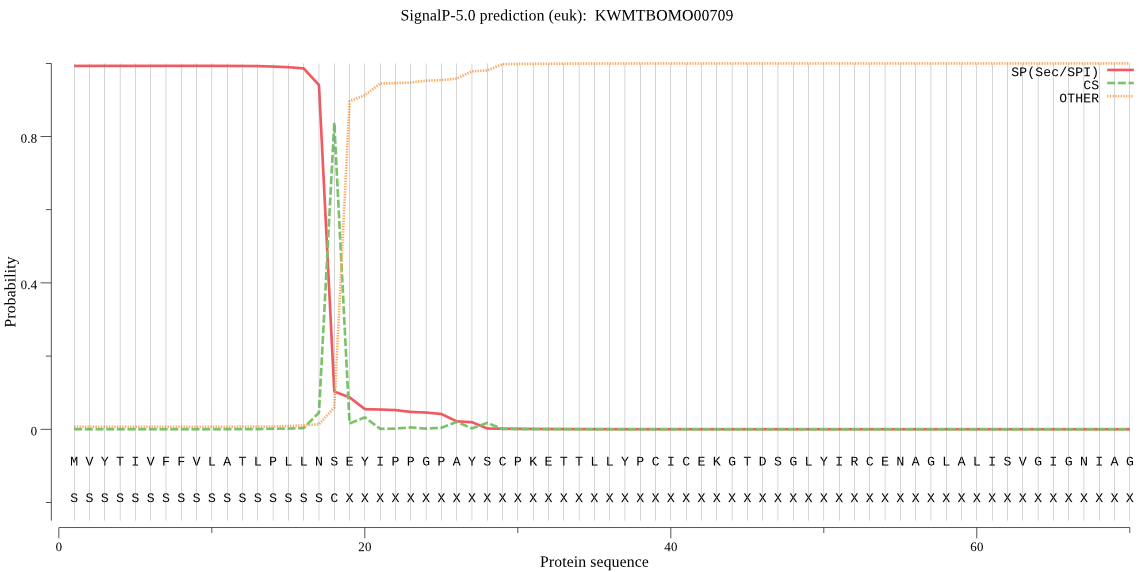
Length:
1117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
20.05764
Exp number, first 60 AAs:
1.44386
Total prob of N-in:
0.13088
outside
1 - 1117
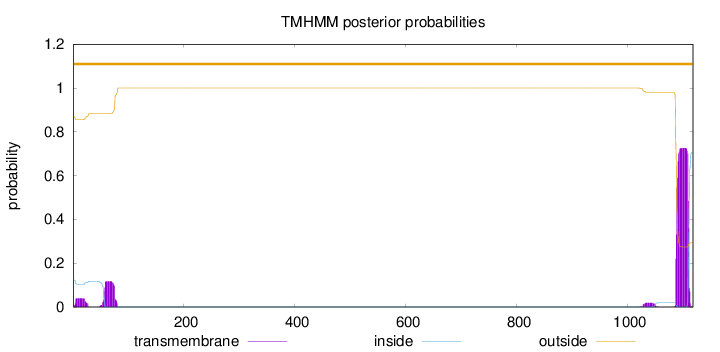
Population Genetic Test Statistics
Pi
213.950348
Theta
159.884124
Tajima's D
1.231795
CLR
0.435486
CSRT
0.71916404179791
Interpretation
Uncertain
