Gene
KWMTBOMO00703
Pre Gene Modal
BGIBMGA003272
Annotation
PREDICTED:_uncharacterized_protein_LOC101746039_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.159
Sequence
CDS
ATGCTGCCGTATAGAAGATTCTGCGGGATGAAACTGAACGTAGGATTCTTGGCCGTATTTCTGTATTTAATAGACAAATCTATACAGCAAGCCATTGATGATTACGTGCCGGGCGAAGACTACCCGGCTTTCACGGAAGTCCCAAAGGGTCTGTCTTTCACCTGCAATGACAAGATACCGGGATATTACGCAGATCCTGAAACCAATTGCCAAGTGTGGCATTGGTGCGTGCCAGGCATCGGTGGCAACCAGCAGTATTCCTTCTTGTGCGGTATCGGGACTGTCTTCAACCAGAGAACGAGGGTCTGCGACTATTTCTTTAAGGTCGACTGTCCGAACTCTCCAGCCTACTACGGAGTCAACGAAGATCTTTACAAAGATGAAGCAGGAAACTACATCAGCGGCAAATAA
Protein
MLPYRRFCGMKLNVGFLAVFLYLIDKSIQQAIDDYVPGEDYPAFTEVPKGLSFTCNDKIPGYYADPETNCQVWHWCVPGIGGNQQYSFLCGIGTVFNQRTRVCDYFFKVDCPNSPAYYGVNEDLYKDEAGNYISGK
Summary
Uniprot
H9J185
A0A2W1BUM1
I4DNR7
A0A0N1INP6
A0A194PUQ7
A0A2A4J5N4
+ More
A0A2H1X1J6 A0A232EVH2 D1MAI0 A0A154PBQ7 A0A026W3V2 A0A151WYV6 E2A1D6 A0A3L8E5K9 A0A195DN96 F4W4J6 A0A195EZJ2 A0A158P339 A0A1L8E389 A0A1L8E3G9 K7J6D1 A0A195BH92 E2C3K4 A0A2A3EN37 A0A222NTA9 A0A087ZS42 E9IIL8 A0A195CCT2 A0A0N0U6X8 A0A0L7RCB4 A0A182MFW7 A0A182FF02 A0A1Y1LDJ3 A0A182NJ60 T1I9N3 A0A182QPK2 A0A182X8Y1 A0A084VCU7 A0A182J959 A0A2R7VXD0 A0A182YP91 A0A182PID7 A0A1A9X0H9 A0A3B0JYQ0 B4IWZ3 W8C3V2 A0A0A1XGZ2 A0A0Q9WVI7 A0A1S4F988 Q9VSE5 A0A034WEP2 A0A1W4VA64 B3DND8 B3DNE4 Q29FD3 B4MXE7 A0A1J1I917 B3NF45 B4PCT7 B3M413 A0A0J9UGU1 M9PEI0 Q17BM7 A0A182LL09 A0A182I413 Q7PP08 A0A182TVW6 B4QLI8 B4HJ03 A0A182VKW0 A0A0L0CEW6 A0A182RMM0 T1JG13 T1L5A2 A0A0K2UXQ1 A0A0P4XWD8 A0A0K2V8M3 A0A0P5AZ03 A0A0P5YCX8 A0A0P5ART6 A0A164XIC7 A0A0P5XBJ2 A0A0N8A4W1
A0A2H1X1J6 A0A232EVH2 D1MAI0 A0A154PBQ7 A0A026W3V2 A0A151WYV6 E2A1D6 A0A3L8E5K9 A0A195DN96 F4W4J6 A0A195EZJ2 A0A158P339 A0A1L8E389 A0A1L8E3G9 K7J6D1 A0A195BH92 E2C3K4 A0A2A3EN37 A0A222NTA9 A0A087ZS42 E9IIL8 A0A195CCT2 A0A0N0U6X8 A0A0L7RCB4 A0A182MFW7 A0A182FF02 A0A1Y1LDJ3 A0A182NJ60 T1I9N3 A0A182QPK2 A0A182X8Y1 A0A084VCU7 A0A182J959 A0A2R7VXD0 A0A182YP91 A0A182PID7 A0A1A9X0H9 A0A3B0JYQ0 B4IWZ3 W8C3V2 A0A0A1XGZ2 A0A0Q9WVI7 A0A1S4F988 Q9VSE5 A0A034WEP2 A0A1W4VA64 B3DND8 B3DNE4 Q29FD3 B4MXE7 A0A1J1I917 B3NF45 B4PCT7 B3M413 A0A0J9UGU1 M9PEI0 Q17BM7 A0A182LL09 A0A182I413 Q7PP08 A0A182TVW6 B4QLI8 B4HJ03 A0A182VKW0 A0A0L0CEW6 A0A182RMM0 T1JG13 T1L5A2 A0A0K2UXQ1 A0A0P4XWD8 A0A0K2V8M3 A0A0P5AZ03 A0A0P5YCX8 A0A0P5ART6 A0A164XIC7 A0A0P5XBJ2 A0A0N8A4W1
Pubmed
19121390
28756777
22651552
26354079
28648823
18362917
+ More
20144715 19820115 24508170 20798317 30249741 21719571 21347285 20075255 28368027 21282665 28004739 24438588 25244985 17994087 24495485 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25348373 15632085 17550304 22936249 17510324 20966253 12364791 14747013 17210077 26108605
20144715 19820115 24508170 20798317 30249741 21719571 21347285 20075255 28368027 21282665 28004739 24438588 25244985 17994087 24495485 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25348373 15632085 17550304 22936249 17510324 20966253 12364791 14747013 17210077 26108605
EMBL
BABH01031446
KZ149912
PZC78061.1
AK403098
BAM19557.1
KQ460940
+ More
KPJ10627.1 KQ459593 KPI96718.1 NWSH01003170 PCG66834.1 ODYU01012706 SOQ59147.1 NNAY01001999 OXU22349.1 GU128088 KQ971354 ACY95471.1 EFA06942.1 KQ434853 KZC08630.1 KK107447 EZA50732.1 KQ982649 KYQ53063.1 GL435760 EFN72762.1 QOIP01000001 RLU27595.1 KQ980713 KYN14301.1 GL887532 EGI70891.1 KQ981905 KYN33546.1 ADTU01007721 GFDF01000881 JAV13203.1 GFDF01000878 JAV13206.1 KQ976467 KYM84139.1 GL452320 EFN77542.1 KZ288204 PBC33148.1 KX503043 ASQ42719.1 GL763494 EFZ19609.1 KQ977935 KYM98669.1 KQ435714 KOX79151.1 KQ414615 KOC68637.1 AXCM01015737 GEZM01058595 JAV71729.1 ACPB03000352 AXCN02000455 ATLV01010912 KE524622 KFB35791.1 KK854102 PTY11340.1 OUUW01000012 SPP87174.1 CH916366 EDV96299.1 GAMC01009956 JAB96599.1 GBXI01004534 JAD09758.1 CH940647 KRF84288.1 AE014296 AAF50478.3 GAKP01006729 JAC52223.1 BT032926 ACD99490.1 BT032932 ACD99496.1 CH379067 EAL31322.3 CH963876 EDW76716.2 CVRI01000043 CRK96052.1 CH954178 EDV50387.2 CM000159 EDW93841.2 CH902618 EDV40375.2 CM002912 KMY98295.1 AGB94258.1 CH477319 EAT43686.1 APCN01002264 AAAB01008960 EAA11225.4 CM000363 EDX09648.1 CH480815 EDW40657.1 JRES01000493 KNC30791.1 JH432192 CAEY01001235 HACA01025683 CDW43044.1 GDIP01236852 JAI86549.1 HACA01029319 CDW46680.1 GDIP01192303 JAJ31099.1 GDIP01059467 JAM44248.1 GDIP01195354 JAJ28048.1 LRGB01000996 KZS14269.1 GDIP01074571 JAM29144.1 GDIP01182249 JAJ41153.1
KPJ10627.1 KQ459593 KPI96718.1 NWSH01003170 PCG66834.1 ODYU01012706 SOQ59147.1 NNAY01001999 OXU22349.1 GU128088 KQ971354 ACY95471.1 EFA06942.1 KQ434853 KZC08630.1 KK107447 EZA50732.1 KQ982649 KYQ53063.1 GL435760 EFN72762.1 QOIP01000001 RLU27595.1 KQ980713 KYN14301.1 GL887532 EGI70891.1 KQ981905 KYN33546.1 ADTU01007721 GFDF01000881 JAV13203.1 GFDF01000878 JAV13206.1 KQ976467 KYM84139.1 GL452320 EFN77542.1 KZ288204 PBC33148.1 KX503043 ASQ42719.1 GL763494 EFZ19609.1 KQ977935 KYM98669.1 KQ435714 KOX79151.1 KQ414615 KOC68637.1 AXCM01015737 GEZM01058595 JAV71729.1 ACPB03000352 AXCN02000455 ATLV01010912 KE524622 KFB35791.1 KK854102 PTY11340.1 OUUW01000012 SPP87174.1 CH916366 EDV96299.1 GAMC01009956 JAB96599.1 GBXI01004534 JAD09758.1 CH940647 KRF84288.1 AE014296 AAF50478.3 GAKP01006729 JAC52223.1 BT032926 ACD99490.1 BT032932 ACD99496.1 CH379067 EAL31322.3 CH963876 EDW76716.2 CVRI01000043 CRK96052.1 CH954178 EDV50387.2 CM000159 EDW93841.2 CH902618 EDV40375.2 CM002912 KMY98295.1 AGB94258.1 CH477319 EAT43686.1 APCN01002264 AAAB01008960 EAA11225.4 CM000363 EDX09648.1 CH480815 EDW40657.1 JRES01000493 KNC30791.1 JH432192 CAEY01001235 HACA01025683 CDW43044.1 GDIP01236852 JAI86549.1 HACA01029319 CDW46680.1 GDIP01192303 JAJ31099.1 GDIP01059467 JAM44248.1 GDIP01195354 JAJ28048.1 LRGB01000996 KZS14269.1 GDIP01074571 JAM29144.1 GDIP01182249 JAJ41153.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000215335
UP000007266
+ More
UP000076502 UP000053097 UP000075809 UP000000311 UP000279307 UP000078492 UP000007755 UP000078541 UP000005205 UP000002358 UP000078540 UP000008237 UP000242457 UP000005203 UP000078542 UP000053105 UP000053825 UP000075883 UP000069272 UP000075884 UP000015103 UP000075886 UP000076407 UP000030765 UP000075880 UP000076408 UP000075885 UP000091820 UP000268350 UP000001070 UP000008792 UP000000803 UP000192221 UP000001819 UP000007798 UP000183832 UP000008711 UP000002282 UP000007801 UP000008820 UP000075882 UP000075840 UP000007062 UP000075902 UP000000304 UP000001292 UP000075903 UP000037069 UP000075900 UP000015104 UP000076858
UP000076502 UP000053097 UP000075809 UP000000311 UP000279307 UP000078492 UP000007755 UP000078541 UP000005205 UP000002358 UP000078540 UP000008237 UP000242457 UP000005203 UP000078542 UP000053105 UP000053825 UP000075883 UP000069272 UP000075884 UP000015103 UP000075886 UP000076407 UP000030765 UP000075880 UP000076408 UP000075885 UP000091820 UP000268350 UP000001070 UP000008792 UP000000803 UP000192221 UP000001819 UP000007798 UP000183832 UP000008711 UP000002282 UP000007801 UP000008820 UP000075882 UP000075840 UP000007062 UP000075902 UP000000304 UP000001292 UP000075903 UP000037069 UP000075900 UP000015104 UP000076858
Pfam
PF01607 CBM_14
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
H9J185
A0A2W1BUM1
I4DNR7
A0A0N1INP6
A0A194PUQ7
A0A2A4J5N4
+ More
A0A2H1X1J6 A0A232EVH2 D1MAI0 A0A154PBQ7 A0A026W3V2 A0A151WYV6 E2A1D6 A0A3L8E5K9 A0A195DN96 F4W4J6 A0A195EZJ2 A0A158P339 A0A1L8E389 A0A1L8E3G9 K7J6D1 A0A195BH92 E2C3K4 A0A2A3EN37 A0A222NTA9 A0A087ZS42 E9IIL8 A0A195CCT2 A0A0N0U6X8 A0A0L7RCB4 A0A182MFW7 A0A182FF02 A0A1Y1LDJ3 A0A182NJ60 T1I9N3 A0A182QPK2 A0A182X8Y1 A0A084VCU7 A0A182J959 A0A2R7VXD0 A0A182YP91 A0A182PID7 A0A1A9X0H9 A0A3B0JYQ0 B4IWZ3 W8C3V2 A0A0A1XGZ2 A0A0Q9WVI7 A0A1S4F988 Q9VSE5 A0A034WEP2 A0A1W4VA64 B3DND8 B3DNE4 Q29FD3 B4MXE7 A0A1J1I917 B3NF45 B4PCT7 B3M413 A0A0J9UGU1 M9PEI0 Q17BM7 A0A182LL09 A0A182I413 Q7PP08 A0A182TVW6 B4QLI8 B4HJ03 A0A182VKW0 A0A0L0CEW6 A0A182RMM0 T1JG13 T1L5A2 A0A0K2UXQ1 A0A0P4XWD8 A0A0K2V8M3 A0A0P5AZ03 A0A0P5YCX8 A0A0P5ART6 A0A164XIC7 A0A0P5XBJ2 A0A0N8A4W1
A0A2H1X1J6 A0A232EVH2 D1MAI0 A0A154PBQ7 A0A026W3V2 A0A151WYV6 E2A1D6 A0A3L8E5K9 A0A195DN96 F4W4J6 A0A195EZJ2 A0A158P339 A0A1L8E389 A0A1L8E3G9 K7J6D1 A0A195BH92 E2C3K4 A0A2A3EN37 A0A222NTA9 A0A087ZS42 E9IIL8 A0A195CCT2 A0A0N0U6X8 A0A0L7RCB4 A0A182MFW7 A0A182FF02 A0A1Y1LDJ3 A0A182NJ60 T1I9N3 A0A182QPK2 A0A182X8Y1 A0A084VCU7 A0A182J959 A0A2R7VXD0 A0A182YP91 A0A182PID7 A0A1A9X0H9 A0A3B0JYQ0 B4IWZ3 W8C3V2 A0A0A1XGZ2 A0A0Q9WVI7 A0A1S4F988 Q9VSE5 A0A034WEP2 A0A1W4VA64 B3DND8 B3DNE4 Q29FD3 B4MXE7 A0A1J1I917 B3NF45 B4PCT7 B3M413 A0A0J9UGU1 M9PEI0 Q17BM7 A0A182LL09 A0A182I413 Q7PP08 A0A182TVW6 B4QLI8 B4HJ03 A0A182VKW0 A0A0L0CEW6 A0A182RMM0 T1JG13 T1L5A2 A0A0K2UXQ1 A0A0P4XWD8 A0A0K2V8M3 A0A0P5AZ03 A0A0P5YCX8 A0A0P5ART6 A0A164XIC7 A0A0P5XBJ2 A0A0N8A4W1
PDB
6G9E
E-value=0.0107023,
Score=84
Ontologies
GO
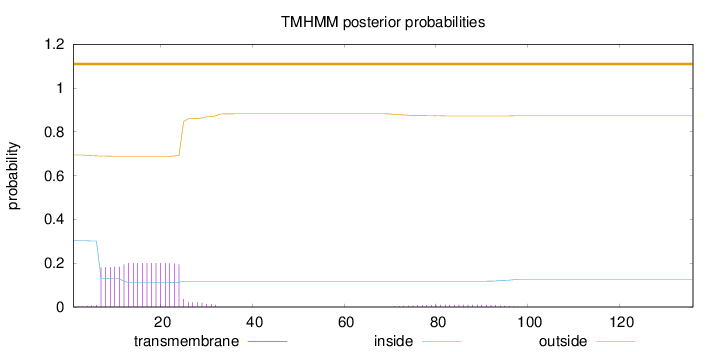
Topology
SignalP
Position: 1 - 30,
Likelihood: 0.713282

Length:
136
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.89898
Exp number, first 60 AAs:
3.6846
Total prob of N-in:
0.30551
outside
1 - 136
Population Genetic Test Statistics
Pi
255.106028
Theta
163.884344
Tajima's D
2.062102
CLR
0
CSRT
0.890555472226389
Interpretation
Uncertain
