Gene
KWMTBOMO00689
Pre Gene Modal
BGIBMGA003222
Annotation
PREDICTED:_BTB/POZ_domain-containing_protein_10_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 3.028
Sequence
CDS
ATGTCTGACTCTCAGGCACATGGTGAGGCAATGAGTGATCGCAGGTCGTTTTTCTATCCTGATAGCAGCAGTGGCTCGGAAGAGTACAACAGAGACGCGGAGGAGCGTCGGAGACGTCTCTCAAAGCGATCTGGAGTATCGAACAGGCCCCGCGCTCCTACAATGCAGTCCGGGGCGTGTTCGTCTAGTAACCCGATGCCATCGACCTCTTCAGCTCAGGAACCTCCGAAAAACGCTCCGAAGCACCAACTCGGAGATGACCGAATCACGTTGGTCGTGGACAATACGAGGTTTGTCGTCGATCCTTCTCAGTTCACGGCGCATCCCAACACAATGCTTGGGAGAATGTTTAGTTCTGGGATAGAATTTACACATCCAAATGAAAGAGGCGAATATGAAGTAGCTGAAGGTATCTCCGCGACCGTATTCAGAGCTATACTGGAATATTACCGAGGAGGTACAATTCGCTGTCCCCCCACGGTGTCTGTGCAAGAGCTGCGAGAGGCCTGTGATTATTTGTTGGTGCCTTTTGATGCTAACACAGTGAGGTGCCAGAACCTCCGTGGCTTATTACATGAGCTATCCAATGAAGGTGCCCGCCGTCAGTTTGAATCGTTCTTGGAGCGTCTCGTACTCCCGCTGATGGTGGAGTCTGCTCGGCGCGGAGACCGCGAGTGTCACGTGGTGGTGTTGCTCGATGATGATGCTGTGGACTGGGACGAGGAATATCCTCCCCAGATGGGAGACGAATACAGTCAGACTGTGCTGTCCACACCGTTGTATAGGTTCTTCAAGTATATTGAGAATCGGGACGTAGCAAAACAAGTAATGAAGGAGAGAGGACTAAAAAAGATAAGGCTTGGCGTCGAAGGGTACCCGACTTACAAGGAGAAAGTTCGCAAACGTCCCGGCGGCAGAGCTGAAGTCATCTACAATTATGTCCAGAGACCGTTCATCCACATGTCCTGGGAGAAGGAGGAAGCAAAGAGCCGCCACGTAGACTTCCAGTGCTTCAAGTCGAAATCCGTCACAAACCTCGCTGAAGCTACAGCGGATCCTGTCATAGAATTAGTGCCGGTAGCGAGGAGAGAAGAAGAAGGTGGAGAAGTGAATGATCCGCCAGAGGAAGAGTGA
Protein
MSDSQAHGEAMSDRRSFFYPDSSSGSEEYNRDAEERRRRLSKRSGVSNRPRAPTMQSGACSSSNPMPSTSSAQEPPKNAPKHQLGDDRITLVVDNTRFVVDPSQFTAHPNTMLGRMFSSGIEFTHPNERGEYEVAEGISATVFRAILEYYRGGTIRCPPTVSVQELREACDYLLVPFDANTVRCQNLRGLLHELSNEGARRQFESFLERLVLPLMVESARRGDRECHVVVLLDDDAVDWDEEYPPQMGDEYSQTVLSTPLYRFFKYIENRDVAKQVMKERGLKKIRLGVEGYPTYKEKVRKRPGGRAEVIYNYVQRPFIHMSWEKEEAKSRHVDFQCFKSKSVTNLAEATADPVIELVPVARREEEGGEVNDPPEEE
Summary
Uniprot
H9J135
A0A0N1IG04
A0A194PTR2
A0A2A4K1L0
A0A212FI73
A0A2W1BSK8
+ More
A0A023F498 A0A069DTY1 A0A0V0G9Y1 A0A0P4VVG4 A0A224XKN1 A0A1B6IB22 A0A1B6GTC4 A0A1B6KQ60 T1IDY7 A0A0C9R0H5 A0A1W4WTH6 A0A195FUI4 E9JD94 A0A232EIS0 K7IMK4 A0A151J422 A0A0C9QAT9 A0A151WIH1 F4WMZ5 A0A146L6N2 A0A026W4S6 A0A088ATW4 A0A154P8V6 A0A0M9A6T4 E2A476 A0A0L7QSA3 N6TSA5 A0A2J7R861 A0A067QML5 A0A139WK37 A0A310SPB0 A0A1W4X326 A0A1W4X495 A0A1Y1MRR8 A0A226E8V0 D6WI26 A0A1Y1MRU1 A0A1Y1MRN0 A0A1L8DAL8 Q0IFH6 A0A1L8DAS8 A0A1Q3FXC6 A0A1B0CNU3 A0A1Q3FXA2 A0A023EPB2 A0A182HA74 E0VZX0 A0A1L8DAQ4 A0A1L8DB58 A0A1Q3FXB0 A0A151HYR8 A0A0P4WHS6 A0A2S2Q4R2 J9JMS8 A0A2H8TRE0 B3M4Y5 B5DQU2 B4H5C4 Q0E8K5 B3NEL2 B4PC23 A0A3B0K2Q5 A0A2M4BLX8 A0A182PEX3 A0A182SLX5 A0A182X247 A0A2M4BLI7 A0A182R9B4 A0A0J9RL36 A0A182I6A8 A0A0A1XD10 A0A182L431 A0A084WN00 Q7QIC3 A0A034W2K2 A0A182LY98 A0A0K8VJK9 A0A182UJT4 A0A182YJH6 A0A182NLN2 W8BMR6 E2BLC3 A0A182VFH5 A0A182WI60 A0A2M4CNX9 A0A2M4AK36 A0A2M4ALK0 A0A2M3ZIR5 A0A182K1V7 A0A0J7L8P6
A0A023F498 A0A069DTY1 A0A0V0G9Y1 A0A0P4VVG4 A0A224XKN1 A0A1B6IB22 A0A1B6GTC4 A0A1B6KQ60 T1IDY7 A0A0C9R0H5 A0A1W4WTH6 A0A195FUI4 E9JD94 A0A232EIS0 K7IMK4 A0A151J422 A0A0C9QAT9 A0A151WIH1 F4WMZ5 A0A146L6N2 A0A026W4S6 A0A088ATW4 A0A154P8V6 A0A0M9A6T4 E2A476 A0A0L7QSA3 N6TSA5 A0A2J7R861 A0A067QML5 A0A139WK37 A0A310SPB0 A0A1W4X326 A0A1W4X495 A0A1Y1MRR8 A0A226E8V0 D6WI26 A0A1Y1MRU1 A0A1Y1MRN0 A0A1L8DAL8 Q0IFH6 A0A1L8DAS8 A0A1Q3FXC6 A0A1B0CNU3 A0A1Q3FXA2 A0A023EPB2 A0A182HA74 E0VZX0 A0A1L8DAQ4 A0A1L8DB58 A0A1Q3FXB0 A0A151HYR8 A0A0P4WHS6 A0A2S2Q4R2 J9JMS8 A0A2H8TRE0 B3M4Y5 B5DQU2 B4H5C4 Q0E8K5 B3NEL2 B4PC23 A0A3B0K2Q5 A0A2M4BLX8 A0A182PEX3 A0A182SLX5 A0A182X247 A0A2M4BLI7 A0A182R9B4 A0A0J9RL36 A0A182I6A8 A0A0A1XD10 A0A182L431 A0A084WN00 Q7QIC3 A0A034W2K2 A0A182LY98 A0A0K8VJK9 A0A182UJT4 A0A182YJH6 A0A182NLN2 W8BMR6 E2BLC3 A0A182VFH5 A0A182WI60 A0A2M4CNX9 A0A2M4AK36 A0A2M4ALK0 A0A2M3ZIR5 A0A182K1V7 A0A0J7L8P6
Pubmed
19121390
26354079
22118469
28756777
25474469
26334808
+ More
27129103 21282665 28648823 20075255 21719571 26823975 24508170 30249741 20798317 23537049 24845553 18362917 19820115 28004739 17510324 24945155 26483478 20566863 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 17550304 22936249 25830018 20966253 24438588 12364791 14747013 17210077 25348373 25244985 24495485
27129103 21282665 28648823 20075255 21719571 26823975 24508170 30249741 20798317 23537049 24845553 18362917 19820115 28004739 17510324 24945155 26483478 20566863 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 17550304 22936249 25830018 20966253 24438588 12364791 14747013 17210077 25348373 25244985 24495485
EMBL
BABH01031464
KQ460940
KPJ10617.1
KQ459593
KPI96707.1
NWSH01000277
+ More
PCG77818.1 AGBW02008426 OWR53431.1 KZ149912 PZC78072.1 GBBI01002367 JAC16345.1 GBGD01001658 JAC87231.1 GECL01001196 JAP04928.1 GDKW01002986 JAI53609.1 GFTR01006048 JAW10378.1 GECU01023646 JAS84060.1 GECZ01004124 JAS65645.1 GEBQ01026420 JAT13557.1 ACPB03000233 GBYB01000311 JAG70078.1 KQ981261 KYN44081.1 GL771866 EFZ09282.1 NNAY01004164 OXU18260.1 KQ980211 KYN17192.1 GBYB01000309 JAG70076.1 KQ983089 KYQ47590.1 GL888223 EGI64465.1 GDHC01014535 JAQ04094.1 KK107419 QOIP01000003 EZA51057.1 RLU25085.1 KQ434846 KZC08283.1 KQ435724 KOX78425.1 GL436596 EFN71747.1 KQ414758 KOC61530.1 APGK01021186 KB740293 KB631672 ENN80943.1 ERL85270.1 NEVH01006723 PNF37027.1 KK853156 KDR10487.1 KQ971334 KYB28264.1 KQ760514 OAD60034.1 GEZM01023495 GEZM01023492 JAV88363.1 LNIX01000006 OXA53291.1 EFA01048.2 GEZM01023489 JAV88369.1 GEZM01023496 JAV88362.1 GFDF01010573 JAV03511.1 CH477317 EAT43738.1 GFDF01010552 JAV03532.1 GFDL01002962 JAV32083.1 AJWK01020902 GFDL01002939 JAV32106.1 GAPW01002286 JAC11312.1 JXUM01122183 JXUM01122184 KQ566576 KXJ70013.1 DS235854 EEB18926.1 GFDF01010531 JAV03553.1 GFDF01010530 JAV03554.1 GFDL01002982 JAV32063.1 KQ976720 KYM76720.1 GDRN01051555 GDRN01051554 JAI66389.1 GGMS01003388 MBY72591.1 ABLF02029343 ABLF02029348 GFXV01004656 MBW16461.1 CH902618 EDV40559.1 CH379070 EDY73975.2 CH479211 EDW32976.1 AE014296 BT099621 AAF47344.1 AAF47345.1 ACU51765.1 CH954178 EDV50007.1 KQS43005.1 CM000159 EDW92678.1 KRK00650.1 OUUW01000012 SPP86942.1 GGFJ01004948 MBW54089.1 GGFJ01004799 MBW53940.1 CM002912 KMY96586.1 APCN01003602 GBXI01005008 JAD09284.1 ATLV01024542 KE525352 KFB51594.1 AAAB01008807 EAA04715.3 GAKP01010028 JAC48924.1 AXCM01000817 GDHF01013261 JAI39053.1 GAMC01004160 JAC02396.1 GL449017 EFN83475.1 GGFL01002713 MBW66891.1 GGFK01007829 MBW41150.1 GGFK01008340 MBW41661.1 GGFM01007651 MBW28402.1 LBMM01000187 KMR04546.1
PCG77818.1 AGBW02008426 OWR53431.1 KZ149912 PZC78072.1 GBBI01002367 JAC16345.1 GBGD01001658 JAC87231.1 GECL01001196 JAP04928.1 GDKW01002986 JAI53609.1 GFTR01006048 JAW10378.1 GECU01023646 JAS84060.1 GECZ01004124 JAS65645.1 GEBQ01026420 JAT13557.1 ACPB03000233 GBYB01000311 JAG70078.1 KQ981261 KYN44081.1 GL771866 EFZ09282.1 NNAY01004164 OXU18260.1 KQ980211 KYN17192.1 GBYB01000309 JAG70076.1 KQ983089 KYQ47590.1 GL888223 EGI64465.1 GDHC01014535 JAQ04094.1 KK107419 QOIP01000003 EZA51057.1 RLU25085.1 KQ434846 KZC08283.1 KQ435724 KOX78425.1 GL436596 EFN71747.1 KQ414758 KOC61530.1 APGK01021186 KB740293 KB631672 ENN80943.1 ERL85270.1 NEVH01006723 PNF37027.1 KK853156 KDR10487.1 KQ971334 KYB28264.1 KQ760514 OAD60034.1 GEZM01023495 GEZM01023492 JAV88363.1 LNIX01000006 OXA53291.1 EFA01048.2 GEZM01023489 JAV88369.1 GEZM01023496 JAV88362.1 GFDF01010573 JAV03511.1 CH477317 EAT43738.1 GFDF01010552 JAV03532.1 GFDL01002962 JAV32083.1 AJWK01020902 GFDL01002939 JAV32106.1 GAPW01002286 JAC11312.1 JXUM01122183 JXUM01122184 KQ566576 KXJ70013.1 DS235854 EEB18926.1 GFDF01010531 JAV03553.1 GFDF01010530 JAV03554.1 GFDL01002982 JAV32063.1 KQ976720 KYM76720.1 GDRN01051555 GDRN01051554 JAI66389.1 GGMS01003388 MBY72591.1 ABLF02029343 ABLF02029348 GFXV01004656 MBW16461.1 CH902618 EDV40559.1 CH379070 EDY73975.2 CH479211 EDW32976.1 AE014296 BT099621 AAF47344.1 AAF47345.1 ACU51765.1 CH954178 EDV50007.1 KQS43005.1 CM000159 EDW92678.1 KRK00650.1 OUUW01000012 SPP86942.1 GGFJ01004948 MBW54089.1 GGFJ01004799 MBW53940.1 CM002912 KMY96586.1 APCN01003602 GBXI01005008 JAD09284.1 ATLV01024542 KE525352 KFB51594.1 AAAB01008807 EAA04715.3 GAKP01010028 JAC48924.1 AXCM01000817 GDHF01013261 JAI39053.1 GAMC01004160 JAC02396.1 GL449017 EFN83475.1 GGFL01002713 MBW66891.1 GGFK01007829 MBW41150.1 GGFK01008340 MBW41661.1 GGFM01007651 MBW28402.1 LBMM01000187 KMR04546.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000015103
+ More
UP000192223 UP000078541 UP000215335 UP000002358 UP000078492 UP000075809 UP000007755 UP000053097 UP000279307 UP000005203 UP000076502 UP000053105 UP000000311 UP000053825 UP000019118 UP000030742 UP000235965 UP000027135 UP000007266 UP000198287 UP000008820 UP000092461 UP000069940 UP000249989 UP000009046 UP000078540 UP000007819 UP000007801 UP000001819 UP000008744 UP000000803 UP000008711 UP000002282 UP000268350 UP000075885 UP000075901 UP000076407 UP000075900 UP000075840 UP000075882 UP000030765 UP000007062 UP000075883 UP000075902 UP000076408 UP000075884 UP000008237 UP000075903 UP000075920 UP000075881 UP000036403
UP000192223 UP000078541 UP000215335 UP000002358 UP000078492 UP000075809 UP000007755 UP000053097 UP000279307 UP000005203 UP000076502 UP000053105 UP000000311 UP000053825 UP000019118 UP000030742 UP000235965 UP000027135 UP000007266 UP000198287 UP000008820 UP000092461 UP000069940 UP000249989 UP000009046 UP000078540 UP000007819 UP000007801 UP000001819 UP000008744 UP000000803 UP000008711 UP000002282 UP000268350 UP000075885 UP000075901 UP000076407 UP000075900 UP000075840 UP000075882 UP000030765 UP000007062 UP000075883 UP000075902 UP000076408 UP000075884 UP000008237 UP000075903 UP000075920 UP000075881 UP000036403
Pfam
Interpro
IPR039885
BTBD10/KCTD20_BTB/POZ
+ More
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
IPR039886 BTBD10/KCTD20
IPR001060 FCH_dom
IPR027267 AH/BAR_dom_sf
IPR031160 F_BAR
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR035460 FCHSD_SH3_1
IPR021896 Transposase_37
IPR011993 PH-like_dom_sf
IPR006612 THAP_Znf
IPR002017 Spectrin_repeat
IPR001849 PH_domain
IPR000219 DH-domain
IPR018159 Spectrin/alpha-actinin
IPR035899 DBL_dom_sf
IPR028570 TRIO
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
IPR039886 BTBD10/KCTD20
IPR001060 FCH_dom
IPR027267 AH/BAR_dom_sf
IPR031160 F_BAR
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR035460 FCHSD_SH3_1
IPR021896 Transposase_37
IPR011993 PH-like_dom_sf
IPR006612 THAP_Znf
IPR002017 Spectrin_repeat
IPR001849 PH_domain
IPR000219 DH-domain
IPR018159 Spectrin/alpha-actinin
IPR035899 DBL_dom_sf
IPR028570 TRIO
Gene 3D
ProteinModelPortal
H9J135
A0A0N1IG04
A0A194PTR2
A0A2A4K1L0
A0A212FI73
A0A2W1BSK8
+ More
A0A023F498 A0A069DTY1 A0A0V0G9Y1 A0A0P4VVG4 A0A224XKN1 A0A1B6IB22 A0A1B6GTC4 A0A1B6KQ60 T1IDY7 A0A0C9R0H5 A0A1W4WTH6 A0A195FUI4 E9JD94 A0A232EIS0 K7IMK4 A0A151J422 A0A0C9QAT9 A0A151WIH1 F4WMZ5 A0A146L6N2 A0A026W4S6 A0A088ATW4 A0A154P8V6 A0A0M9A6T4 E2A476 A0A0L7QSA3 N6TSA5 A0A2J7R861 A0A067QML5 A0A139WK37 A0A310SPB0 A0A1W4X326 A0A1W4X495 A0A1Y1MRR8 A0A226E8V0 D6WI26 A0A1Y1MRU1 A0A1Y1MRN0 A0A1L8DAL8 Q0IFH6 A0A1L8DAS8 A0A1Q3FXC6 A0A1B0CNU3 A0A1Q3FXA2 A0A023EPB2 A0A182HA74 E0VZX0 A0A1L8DAQ4 A0A1L8DB58 A0A1Q3FXB0 A0A151HYR8 A0A0P4WHS6 A0A2S2Q4R2 J9JMS8 A0A2H8TRE0 B3M4Y5 B5DQU2 B4H5C4 Q0E8K5 B3NEL2 B4PC23 A0A3B0K2Q5 A0A2M4BLX8 A0A182PEX3 A0A182SLX5 A0A182X247 A0A2M4BLI7 A0A182R9B4 A0A0J9RL36 A0A182I6A8 A0A0A1XD10 A0A182L431 A0A084WN00 Q7QIC3 A0A034W2K2 A0A182LY98 A0A0K8VJK9 A0A182UJT4 A0A182YJH6 A0A182NLN2 W8BMR6 E2BLC3 A0A182VFH5 A0A182WI60 A0A2M4CNX9 A0A2M4AK36 A0A2M4ALK0 A0A2M3ZIR5 A0A182K1V7 A0A0J7L8P6
A0A023F498 A0A069DTY1 A0A0V0G9Y1 A0A0P4VVG4 A0A224XKN1 A0A1B6IB22 A0A1B6GTC4 A0A1B6KQ60 T1IDY7 A0A0C9R0H5 A0A1W4WTH6 A0A195FUI4 E9JD94 A0A232EIS0 K7IMK4 A0A151J422 A0A0C9QAT9 A0A151WIH1 F4WMZ5 A0A146L6N2 A0A026W4S6 A0A088ATW4 A0A154P8V6 A0A0M9A6T4 E2A476 A0A0L7QSA3 N6TSA5 A0A2J7R861 A0A067QML5 A0A139WK37 A0A310SPB0 A0A1W4X326 A0A1W4X495 A0A1Y1MRR8 A0A226E8V0 D6WI26 A0A1Y1MRU1 A0A1Y1MRN0 A0A1L8DAL8 Q0IFH6 A0A1L8DAS8 A0A1Q3FXC6 A0A1B0CNU3 A0A1Q3FXA2 A0A023EPB2 A0A182HA74 E0VZX0 A0A1L8DAQ4 A0A1L8DB58 A0A1Q3FXB0 A0A151HYR8 A0A0P4WHS6 A0A2S2Q4R2 J9JMS8 A0A2H8TRE0 B3M4Y5 B5DQU2 B4H5C4 Q0E8K5 B3NEL2 B4PC23 A0A3B0K2Q5 A0A2M4BLX8 A0A182PEX3 A0A182SLX5 A0A182X247 A0A2M4BLI7 A0A182R9B4 A0A0J9RL36 A0A182I6A8 A0A0A1XD10 A0A182L431 A0A084WN00 Q7QIC3 A0A034W2K2 A0A182LY98 A0A0K8VJK9 A0A182UJT4 A0A182YJH6 A0A182NLN2 W8BMR6 E2BLC3 A0A182VFH5 A0A182WI60 A0A2M4CNX9 A0A2M4AK36 A0A2M4ALK0 A0A2M3ZIR5 A0A182K1V7 A0A0J7L8P6
Ontologies
GO
PANTHER
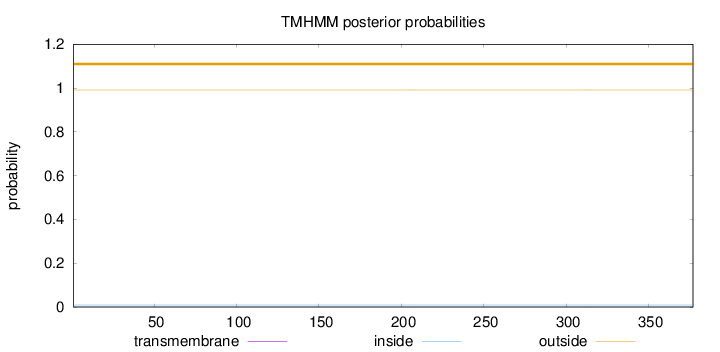
Topology
Length:
377
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00922
outside
1 - 377
Population Genetic Test Statistics
Pi
277.310663
Theta
170.014824
Tajima's D
1.826141
CLR
0.36006
CSRT
0.855207239638018
Interpretation
Uncertain
