Gene
KWMTBOMO00687 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003221
Annotation
PREDICTED:_probable_nucleolar_GTP-binding_protein_1_[Bombyx_mori]
Full name
Nucleolar GTP-binding protein 1
Alternative Name
Novel nucleolar protein 1
Location in the cell
Mitochondrial Reliability : 2.195 Nuclear Reliability : 1.728
Sequence
CDS
ATGAGTTTGTACAACTTTAAAAAGATTGCTGTAGTCCCTACAGCTAAGGACTTTATAGACATAATTTTATCGAAAACACAAAGGAAAACTCCTACAGTCGTTCACAAGCATTATAAAATATCAAGAATACGTGGGTTTTATATTAGAAAAGTGAAATATACCCAGCAGAACTTCCACGACCGACTGTCCAGAATTATTCAGGAATTCCCAAAGTTAGATGATGTCCATCCATTTTATGCTGATCTCATGAACGTATTGTACGACAAGGATCATTACAAACTTGGACTCGGACAGCTGAATACAGCCAGACATCTTATTGACAATGTCGCAAAAGACTATGTGAGACTGTTAAAATATGGAGATTCGTTGTATCGGTGCAAACAGCTCAAACGTGCGGCTCTGGGTCGCATGGCCACCATCATGAAACGGCAAGGAGCTAATCTTACATACCTGGAACAGGTCCGTCAACATTTAGCACGTTTACCATCGATAGATCCCTACACCAGGACGATAATCATCTGTGGATTCCCAAATGTCGGAAAATCAAGTTTTATTAATAAAATAACACGTGCTGATGTGGAAGTGCAGCCATATGCGTTTACCACAAAGAGTCTGTATGTTGGTCATACTGATTACAAATATTTGAGATGGCAGGTGATAGACACCCCTGGAATCTTGGATCATCCATTGGAAGAACGCAATGTTATAGAAATGCAAGCGGTCACAGCTTTGGCTCATCTCAGAGCTGCCATACTATACTTCATAGATCCGTCTGAGCAGTGTGGCCATAGTATTGAAGACCAGATATCACTGTTCGAAAGTATAAAGCCTTTATTCACAAGCAAACCCCTAATAGTAGTGTTAAACAAGATGGATGTGGTCAAACCTGAAGAGCTGTCTCTGAGTAAGAAACAGTTGCTGGACGATCTGACCGAAACGTGCTCTAAAGGAAATCTTGTCAATGCGGATTCAAATTCTGATCTGTCCGTGGTGCCTGTGATGCGGATGTCCACTGTGACTGAAGAGGGCGTGCAGGAGGTCAAAATTGAAGCCTGCGAGAGGCTGCTGGGACACCGGGTCACAGAGAAAATGCGGACAAAGAAGGTGGACGGCATCTTGAACCGGCTCCACGTGGCGGTGCCGGCCCCGCGCGACGACAAGCCGCGCCCGCCCGTGCTGCCGCCCTCGGTCCTCGCCAAGAAGGAGAAACAAGCAGAGAAGGAGAACCGGAAGAGGAAACTGGAGAGGGACATCGAGGTGGAGCTCGGCGACGACTACGTACTCGACTTGAAGAAGAACTACACGGAGATCCCGGAGGACGAGCGCTATGACGTCATCCCGGAGATCTGGGAGGGACACAACATCGCCGACTATATCGACCCGGACATCTTTGATAAACTGGCAGAGTTGGAGAAGGACGAGGAACTGCGCGAGGCTGGCGGCATGTACGCCGCGCCCAAGATCGAGCTGGACGACACCGTGAGGGAGATCAGGGAACTGGCCCGGCAGATACGCAACAAGAAGGCCATTCTCAAGGACGAGTCTCGCCTGGTGAAGCAGTCCACGAAGCCGGTGATGCCGCGCACCTCGCGAGCCAAGACCAAGCAGAGGTCCACCTCCAAGCTCAGGAAGGACATGGAGAAACTTGGCGTGGACATGTCGGAGACCGGCGACGCGCACTTCACCCGCGCCCGCCCCCGCTCCCGCTCCCGCTCGCTCTCCGCGCCGCCCGCCAAGCGCGCGCGGGAACCCTCCAACCAGCCGGTGGCCGACCCCATCATGCGCGTGAAAGTAAAGAGGATGGCGCACGCGGCCATCGCCAAGAAGACCAAGAAGATGGGCCTCAAGGGAGAGGCGGACCGGTTCATCGGCACCAAGATGCCGAAGCATCTGTTCGCTGGAAAACGTGGGGTCGGCAAGACGGACCGGAGATAG
Protein
MSLYNFKKIAVVPTAKDFIDIILSKTQRKTPTVVHKHYKISRIRGFYIRKVKYTQQNFHDRLSRIIQEFPKLDDVHPFYADLMNVLYDKDHYKLGLGQLNTARHLIDNVAKDYVRLLKYGDSLYRCKQLKRAALGRMATIMKRQGANLTYLEQVRQHLARLPSIDPYTRTIIICGFPNVGKSSFINKITRADVEVQPYAFTTKSLYVGHTDYKYLRWQVIDTPGILDHPLEERNVIEMQAVTALAHLRAAILYFIDPSEQCGHSIEDQISLFESIKPLFTSKPLIVVLNKMDVVKPEELSLSKKQLLDDLTETCSKGNLVNADSNSDLSVVPVMRMSTVTEEGVQEVKIEACERLLGHRVTEKMRTKKVDGILNRLHVAVPAPRDDKPRPPVLPPSVLAKKEKQAEKENRKRKLERDIEVELGDDYVLDLKKNYTEIPEDERYDVIPEIWEGHNIADYIDPDIFDKLAELEKDEELREAGGMYAAPKIELDDTVREIRELARQIRNKKAILKDESRLVKQSTKPVMPRTSRAKTKQRSTSKLRKDMEKLGVDMSETGDAHFTRARPRSRSRSLSAPPAKRAREPSNQPVADPIMRVKVKRMAHAAIAKKTKKMGLKGEADRFIGTKMPKHLFAGKRGVGKTDRR
Summary
Description
Involved in the biogenesis of the 60S ribosomal subunit.
Involved in the biogenesis of the 60S ribosomal subunit (By similarity). Required for normal assembly of the mitotic spindle (PubMed:24255106). May be involved in both centrosome-dependent and centrosome-independent spindle assembly programs (PubMed:24255106).
Involved in the biogenesis of the 60S ribosomal subunit (By similarity). Required for normal assembly of the mitotic spindle (PubMed:24255106). May be involved in both centrosome-dependent and centrosome-independent spindle assembly programs (PubMed:24255106).
Similarity
Belongs to the TRAFAC class OBG-HflX-like GTPase superfamily. OBG GTPase family. NOG subfamily.
Keywords
Cell cycle
Cell division
Complete proteome
GTP-binding
Mitosis
Nucleotide-binding
Nucleus
Reference proteome
Ribosome biogenesis
Feature
chain Nucleolar GTP-binding protein 1
Uniprot
A0A2W1BSM8
A0A2A4JB68
A0A194PZV5
D6WR57
A0A1Y1L5U0
A0A1W4XGU9
+ More
J3JWL5 A0A0K8UAU0 A0A088AUU3 A0A0L7QL59 A0A2J7PD57 A0A0A1X0F2 B4LN40 W8BBX4 A0A034WSY7 A0A0K8TRI1 B4KTH4 A0A1W4UYD6 A0A1L8ED98 A0A1L8EDA9 A0A1I8PMP4 A0A2P8YT71 B4P415 B3N7H3 A0A1L8EDI7 E2BWL7 B4MJN7 A0A0B4LEY9 Q9V411 A0A232EG88 K7J2V8 B4J4E9 T1PGH7 B4QH85 B4HSR8 A0A1B6HJI0 A0A0M4EV72 A0A023FCH1 A0A154PK65 A0A1B0FR70 A0A2A3E7Z3 A0A0J7NHX3 A0A1A9VN72 B4GHS6 A0A1B6L217 Q28ZG8 A0A0C9QUM2 D3TPK2 A0A026W2X2 A0A067QRT9 B3MFK3 A0A310SUI6 A0A3B0JCM0 A0A0V0G4Q4 R4WJM9 A0A069DW17 A0A224XKY7 A0A0P4VM80 A0A0L0CPG3 A0A0K8V0Z4 A0A1B0CKD3 F4WL71 A0A0N0BG08 A0A158NZJ0 E2AQY2 A0A1J1ITM5 A0A1L8DHY1 A0A2S2R2Z9 A0A1S4FNZ2 Q16UL5 A0A182MB80 A0A023EUN8 J9JX20 A0A182WL15 A0A182RMX9 A0A1Q3FWH5 A0A182UQ05 Q7QIT0 A0A182HWF5 A0A2M3YXK1 A0A2M3ZZ51 A0A2M4BGM6 A0A182WWS5 T1DQ49 A0A2M4A161 W5JW60 A0A195CIB3 A0A084WJV0 A0A182SD60 A0A0A9ZEK9 C5WLT8 A0A336JY94 A0A182QK04 A0A195BMS3 A0A182IK65 A0A182NJP6 A0A195FCB0 T1ICJ4 A0A182KD56 A0A182Y270
J3JWL5 A0A0K8UAU0 A0A088AUU3 A0A0L7QL59 A0A2J7PD57 A0A0A1X0F2 B4LN40 W8BBX4 A0A034WSY7 A0A0K8TRI1 B4KTH4 A0A1W4UYD6 A0A1L8ED98 A0A1L8EDA9 A0A1I8PMP4 A0A2P8YT71 B4P415 B3N7H3 A0A1L8EDI7 E2BWL7 B4MJN7 A0A0B4LEY9 Q9V411 A0A232EG88 K7J2V8 B4J4E9 T1PGH7 B4QH85 B4HSR8 A0A1B6HJI0 A0A0M4EV72 A0A023FCH1 A0A154PK65 A0A1B0FR70 A0A2A3E7Z3 A0A0J7NHX3 A0A1A9VN72 B4GHS6 A0A1B6L217 Q28ZG8 A0A0C9QUM2 D3TPK2 A0A026W2X2 A0A067QRT9 B3MFK3 A0A310SUI6 A0A3B0JCM0 A0A0V0G4Q4 R4WJM9 A0A069DW17 A0A224XKY7 A0A0P4VM80 A0A0L0CPG3 A0A0K8V0Z4 A0A1B0CKD3 F4WL71 A0A0N0BG08 A0A158NZJ0 E2AQY2 A0A1J1ITM5 A0A1L8DHY1 A0A2S2R2Z9 A0A1S4FNZ2 Q16UL5 A0A182MB80 A0A023EUN8 J9JX20 A0A182WL15 A0A182RMX9 A0A1Q3FWH5 A0A182UQ05 Q7QIT0 A0A182HWF5 A0A2M3YXK1 A0A2M3ZZ51 A0A2M4BGM6 A0A182WWS5 T1DQ49 A0A2M4A161 W5JW60 A0A195CIB3 A0A084WJV0 A0A182SD60 A0A0A9ZEK9 C5WLT8 A0A336JY94 A0A182QK04 A0A195BMS3 A0A182IK65 A0A182NJP6 A0A195FCB0 T1ICJ4 A0A182KD56 A0A182Y270
Pubmed
28756777
26354079
18362917
19820115
28004739
22516182
+ More
25830018 17994087 24495485 25348373 26369729 29403074 17550304 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 24255106 28648823 20075255 25315136 22936249 25474469 15632085 20353571 24508170 30249741 24845553 23691247 26334808 27129103 26108605 21719571 21347285 17510324 24945155 26483478 12364791 14747013 17210077 20920257 23761445 24438588 25401762 26823975 25244985
25830018 17994087 24495485 25348373 26369729 29403074 17550304 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 24255106 28648823 20075255 25315136 22936249 25474469 15632085 20353571 24508170 30249741 24845553 23691247 26334808 27129103 26108605 21719571 21347285 17510324 24945155 26483478 12364791 14747013 17210077 20920257 23761445 24438588 25401762 26823975 25244985
EMBL
KZ149912
PZC78082.1
NWSH01002193
PCG68938.1
KQ459593
KPI96700.1
+ More
KQ971354 EFA06007.1 GEZM01063645 JAV69042.1 BT127633 AEE62595.1 GDHF01028580 GDHF01019997 JAI23734.1 JAI32317.1 KQ414934 KOC59347.1 NEVH01026393 PNF14270.1 GBXI01009946 JAD04346.1 CH940648 EDW60044.1 GAMC01007805 JAB98750.1 GAKP01000286 JAC58666.1 GDAI01000860 JAI16743.1 CH933808 EDW08535.1 GFDG01002092 JAV16707.1 GFDG01002091 JAV16708.1 PYGN01000374 PSN47441.1 CM000157 EDW89498.1 CH954177 EDV59378.1 GFDG01002140 JAV16659.1 GL451131 EFN79973.1 CH963846 EDW72326.1 AE013599 AHN56045.1 AF181654 NNAY01004854 OXU17338.1 CH916367 EDW01631.1 KA647245 AFP61874.1 CM000362 CM002911 EDX06339.1 KMY92505.1 CH480816 EDW47095.1 GECU01032850 JAS74856.1 CP012524 ALC41725.1 GBBI01000239 JAC18473.1 KQ434926 KZC11688.1 CCAG010022994 KZ288356 PBC27171.1 LBMM01004791 KMQ92085.1 CH479183 EDW36046.1 GEBQ01022230 JAT17747.1 CM000071 EAL25645.1 GBYB01007454 JAG77221.1 EZ423354 ADD19630.1 KK107488 QOIP01000012 EZA49936.1 RLU15738.1 KK853011 KDR12537.1 CH902619 EDV36688.1 KQ759866 OAD62489.1 OUUW01000001 SPP72960.1 GECL01003102 JAP03022.1 AK417775 BAN20990.1 GBGD01000779 JAC88110.1 GFTR01007623 JAW08803.1 GDKW01000365 JAI56230.1 JRES01000204 KNC33324.1 GDHF01026847 GDHF01020079 JAI25467.1 JAI32235.1 AJWK01016025 GL888207 EGI65034.1 KQ435794 KOX74112.1 ADTU01004645 GL441846 EFN64128.1 CVRI01000059 CRL03074.1 GFDF01008099 JAV05985.1 GGMS01015173 MBY84376.1 CH477618 EAT38228.1 AXCM01002461 JXUM01002670 GAPW01000458 KQ560143 JAC13140.1 KXJ84199.1 ABLF02032387 GFDL01003108 JAV31937.1 AAAB01008807 EAA03933.2 APCN01001489 GGFM01000239 MBW20990.1 GGFK01000513 MBW33834.1 GGFJ01003064 MBW52205.1 GAMD01002764 JAA98826.1 GGFK01001057 MBW34378.1 ADMH02000263 ETN67194.1 KQ977720 KYN00470.1 ATLV01024074 KE525348 KFB50494.1 GBHO01001811 GBHO01001810 GDHC01020762 JAG41793.1 JAG41794.1 JAP97866.1 BT088903 ACT09400.1 UFQS01000016 UFQT01000016 SSW97419.1 SSX17805.1 AXCN02000497 KQ976433 KYM87288.1 KQ981685 KYN38031.1 ACPB03020780
KQ971354 EFA06007.1 GEZM01063645 JAV69042.1 BT127633 AEE62595.1 GDHF01028580 GDHF01019997 JAI23734.1 JAI32317.1 KQ414934 KOC59347.1 NEVH01026393 PNF14270.1 GBXI01009946 JAD04346.1 CH940648 EDW60044.1 GAMC01007805 JAB98750.1 GAKP01000286 JAC58666.1 GDAI01000860 JAI16743.1 CH933808 EDW08535.1 GFDG01002092 JAV16707.1 GFDG01002091 JAV16708.1 PYGN01000374 PSN47441.1 CM000157 EDW89498.1 CH954177 EDV59378.1 GFDG01002140 JAV16659.1 GL451131 EFN79973.1 CH963846 EDW72326.1 AE013599 AHN56045.1 AF181654 NNAY01004854 OXU17338.1 CH916367 EDW01631.1 KA647245 AFP61874.1 CM000362 CM002911 EDX06339.1 KMY92505.1 CH480816 EDW47095.1 GECU01032850 JAS74856.1 CP012524 ALC41725.1 GBBI01000239 JAC18473.1 KQ434926 KZC11688.1 CCAG010022994 KZ288356 PBC27171.1 LBMM01004791 KMQ92085.1 CH479183 EDW36046.1 GEBQ01022230 JAT17747.1 CM000071 EAL25645.1 GBYB01007454 JAG77221.1 EZ423354 ADD19630.1 KK107488 QOIP01000012 EZA49936.1 RLU15738.1 KK853011 KDR12537.1 CH902619 EDV36688.1 KQ759866 OAD62489.1 OUUW01000001 SPP72960.1 GECL01003102 JAP03022.1 AK417775 BAN20990.1 GBGD01000779 JAC88110.1 GFTR01007623 JAW08803.1 GDKW01000365 JAI56230.1 JRES01000204 KNC33324.1 GDHF01026847 GDHF01020079 JAI25467.1 JAI32235.1 AJWK01016025 GL888207 EGI65034.1 KQ435794 KOX74112.1 ADTU01004645 GL441846 EFN64128.1 CVRI01000059 CRL03074.1 GFDF01008099 JAV05985.1 GGMS01015173 MBY84376.1 CH477618 EAT38228.1 AXCM01002461 JXUM01002670 GAPW01000458 KQ560143 JAC13140.1 KXJ84199.1 ABLF02032387 GFDL01003108 JAV31937.1 AAAB01008807 EAA03933.2 APCN01001489 GGFM01000239 MBW20990.1 GGFK01000513 MBW33834.1 GGFJ01003064 MBW52205.1 GAMD01002764 JAA98826.1 GGFK01001057 MBW34378.1 ADMH02000263 ETN67194.1 KQ977720 KYN00470.1 ATLV01024074 KE525348 KFB50494.1 GBHO01001811 GBHO01001810 GDHC01020762 JAG41793.1 JAG41794.1 JAP97866.1 BT088903 ACT09400.1 UFQS01000016 UFQT01000016 SSW97419.1 SSX17805.1 AXCN02000497 KQ976433 KYM87288.1 KQ981685 KYN38031.1 ACPB03020780
Proteomes
UP000218220
UP000053268
UP000007266
UP000192223
UP000005203
UP000053825
+ More
UP000235965 UP000008792 UP000009192 UP000192221 UP000095300 UP000245037 UP000002282 UP000008711 UP000008237 UP000007798 UP000000803 UP000215335 UP000002358 UP000001070 UP000095301 UP000000304 UP000001292 UP000092553 UP000076502 UP000092444 UP000242457 UP000036403 UP000078200 UP000008744 UP000001819 UP000053097 UP000279307 UP000027135 UP000007801 UP000268350 UP000037069 UP000092461 UP000007755 UP000053105 UP000005205 UP000000311 UP000183832 UP000008820 UP000075883 UP000069940 UP000249989 UP000007819 UP000075920 UP000075900 UP000075903 UP000007062 UP000075840 UP000076407 UP000000673 UP000078542 UP000030765 UP000075901 UP000075886 UP000078540 UP000075880 UP000075884 UP000078541 UP000015103 UP000075881 UP000076408
UP000235965 UP000008792 UP000009192 UP000192221 UP000095300 UP000245037 UP000002282 UP000008711 UP000008237 UP000007798 UP000000803 UP000215335 UP000002358 UP000001070 UP000095301 UP000000304 UP000001292 UP000092553 UP000076502 UP000092444 UP000242457 UP000036403 UP000078200 UP000008744 UP000001819 UP000053097 UP000279307 UP000027135 UP000007801 UP000268350 UP000037069 UP000092461 UP000007755 UP000053105 UP000005205 UP000000311 UP000183832 UP000008820 UP000075883 UP000069940 UP000249989 UP000007819 UP000075920 UP000075900 UP000075903 UP000007062 UP000075840 UP000076407 UP000000673 UP000078542 UP000030765 UP000075901 UP000075886 UP000078540 UP000075880 UP000075884 UP000078541 UP000015103 UP000075881 UP000076408
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BSM8
A0A2A4JB68
A0A194PZV5
D6WR57
A0A1Y1L5U0
A0A1W4XGU9
+ More
J3JWL5 A0A0K8UAU0 A0A088AUU3 A0A0L7QL59 A0A2J7PD57 A0A0A1X0F2 B4LN40 W8BBX4 A0A034WSY7 A0A0K8TRI1 B4KTH4 A0A1W4UYD6 A0A1L8ED98 A0A1L8EDA9 A0A1I8PMP4 A0A2P8YT71 B4P415 B3N7H3 A0A1L8EDI7 E2BWL7 B4MJN7 A0A0B4LEY9 Q9V411 A0A232EG88 K7J2V8 B4J4E9 T1PGH7 B4QH85 B4HSR8 A0A1B6HJI0 A0A0M4EV72 A0A023FCH1 A0A154PK65 A0A1B0FR70 A0A2A3E7Z3 A0A0J7NHX3 A0A1A9VN72 B4GHS6 A0A1B6L217 Q28ZG8 A0A0C9QUM2 D3TPK2 A0A026W2X2 A0A067QRT9 B3MFK3 A0A310SUI6 A0A3B0JCM0 A0A0V0G4Q4 R4WJM9 A0A069DW17 A0A224XKY7 A0A0P4VM80 A0A0L0CPG3 A0A0K8V0Z4 A0A1B0CKD3 F4WL71 A0A0N0BG08 A0A158NZJ0 E2AQY2 A0A1J1ITM5 A0A1L8DHY1 A0A2S2R2Z9 A0A1S4FNZ2 Q16UL5 A0A182MB80 A0A023EUN8 J9JX20 A0A182WL15 A0A182RMX9 A0A1Q3FWH5 A0A182UQ05 Q7QIT0 A0A182HWF5 A0A2M3YXK1 A0A2M3ZZ51 A0A2M4BGM6 A0A182WWS5 T1DQ49 A0A2M4A161 W5JW60 A0A195CIB3 A0A084WJV0 A0A182SD60 A0A0A9ZEK9 C5WLT8 A0A336JY94 A0A182QK04 A0A195BMS3 A0A182IK65 A0A182NJP6 A0A195FCB0 T1ICJ4 A0A182KD56 A0A182Y270
J3JWL5 A0A0K8UAU0 A0A088AUU3 A0A0L7QL59 A0A2J7PD57 A0A0A1X0F2 B4LN40 W8BBX4 A0A034WSY7 A0A0K8TRI1 B4KTH4 A0A1W4UYD6 A0A1L8ED98 A0A1L8EDA9 A0A1I8PMP4 A0A2P8YT71 B4P415 B3N7H3 A0A1L8EDI7 E2BWL7 B4MJN7 A0A0B4LEY9 Q9V411 A0A232EG88 K7J2V8 B4J4E9 T1PGH7 B4QH85 B4HSR8 A0A1B6HJI0 A0A0M4EV72 A0A023FCH1 A0A154PK65 A0A1B0FR70 A0A2A3E7Z3 A0A0J7NHX3 A0A1A9VN72 B4GHS6 A0A1B6L217 Q28ZG8 A0A0C9QUM2 D3TPK2 A0A026W2X2 A0A067QRT9 B3MFK3 A0A310SUI6 A0A3B0JCM0 A0A0V0G4Q4 R4WJM9 A0A069DW17 A0A224XKY7 A0A0P4VM80 A0A0L0CPG3 A0A0K8V0Z4 A0A1B0CKD3 F4WL71 A0A0N0BG08 A0A158NZJ0 E2AQY2 A0A1J1ITM5 A0A1L8DHY1 A0A2S2R2Z9 A0A1S4FNZ2 Q16UL5 A0A182MB80 A0A023EUN8 J9JX20 A0A182WL15 A0A182RMX9 A0A1Q3FWH5 A0A182UQ05 Q7QIT0 A0A182HWF5 A0A2M3YXK1 A0A2M3ZZ51 A0A2M4BGM6 A0A182WWS5 T1DQ49 A0A2M4A161 W5JW60 A0A195CIB3 A0A084WJV0 A0A182SD60 A0A0A9ZEK9 C5WLT8 A0A336JY94 A0A182QK04 A0A195BMS3 A0A182IK65 A0A182NJP6 A0A195FCB0 T1ICJ4 A0A182KD56 A0A182Y270
PDB
6N8L
E-value=1.38559e-142,
Score=1300
Ontologies
KEGG
GO
PANTHER
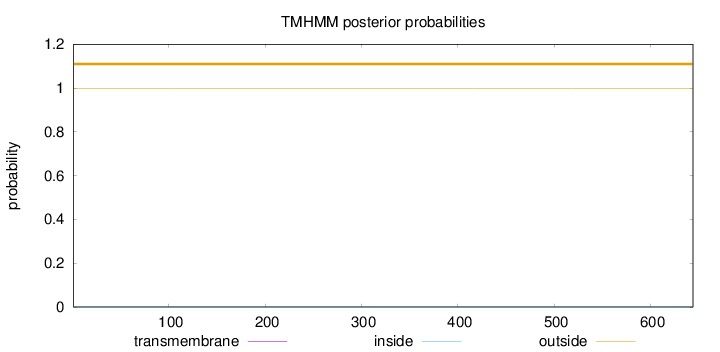
Topology
Subcellular location
Length:
644
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000600000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00086
outside
1 - 644
Population Genetic Test Statistics
Pi
297.678646
Theta
183.645962
Tajima's D
1.882738
CLR
0.005246
CSRT
0.863506824658767
Interpretation
Uncertain
