Gene
KWMTBOMO00686 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003279
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_presequence_protease?_mitochondrial-like_[Bombyx_mori]
Full name
Presequence protease, mitochondrial
Location in the cell
Nuclear Reliability : 2.798
Sequence
CDS
ATGTATTCAAGACTGCGCAATGGTTTTCGGCAAACAGTTAAACACCGACTGGCTTCGCAAAGAAGTTACAGTAATGAGGTTTTGAGGAAAAAGGAAAATTTATCAAAAAATCTTCAAGTTGGGAAGACTATACATGGATTCCTTTGCACCGAAATCGAATCTATACAAGAATACAATATGACAGCTTATTTTTTAACCCACGAGAGAACCAAAACGGAATATTTGCATTTAGAGAGAGATGACTCAAATAATGTATTTTCTGTAGGTTTTCGCACAACTCCGTTAGATTCAATGGGCACTCCCCACATTCTAGAACACACAGTTTTATGCGGCTCAGAAAAATATCCAGTAAGAGATCCATTCTTCAAAATGCTGAACAGATCTTTGGCTACGTTCATGAATGCTCTAACAGGCCCAGATTATACATTCTATCCTTTTTCATCGCAAAATGAAATTGACTATCGTAATCTACAGAAAGTTTATTTGGATGCAGTCTTCAGACCTAATTTAACTAGACTAGATTTTTTACAAGAAGGCTGGAGATTAGAACATTCAAATTTGGAGGATAAAAATTCTGATTTGATTTTCAAAGGTGTAGTCTACAATGAAATGAAGGGAGCCTTTTCGGAGACAAGTTCGCTTTTTGGACAAAAATTCATTAATACAATTTTACCGAAAGGAACATATGGATATGTATCAGGAGGTGACCCGTTATGCATTCCTCAGCTAACTCATGAGCATTTAAAAAACTTCCATGCCACTTATTATCATCCTAGCAATTCAAGAATGTATTCGTATGGCAGTTTTCCATTAGAAGCCAACTTAAAGTTCATTAATGAAGCCTATCTGAGTAAATATGAATATTTAGATCCCAAGAACACTGTAGTTGCGAGTCAAGAACGTTGGAAGGAGCCAAGACGTGCAGAAATGAACTGCAGAGTAGACCAGTATGGTGGACCAGTAGAAAAACAGAATCAAATTGCCATAGGATATGTTATGTCGGACATTACTAATATTTATGAAACATTCACAATGCTAGCATTAGCGGAACTTATGATAATAGGACCGAATTCAGCATTTTACAAAAGCTTAATTGAGAAAAACATATCGGGAGGCTATAATTCTTTGACTGGCTATGAAAATCAGATAAGAGATACCATGTTTGTTGTTGGTCTCCGAGATGTTGAAACGTCAAAGTTTGAGACCATCGAGAAGATTGCAAATCAAACTTTAGAGGAAATATATGAGAAGGGCTTTGAGAAAGCTCATATTGAGAGTGTTTTACATGGCTTCGAGCTTTCTATTAAGCACCAATCACCAAAGTTTGGTTTAAATCTGCTTTTTAACCTAATGCCTCTCTGGAATCACAATGGGCCGATATTGAATGCTTTGAAGGTCAACAAACTTTTGGGGAGACTGAAGGAGAATCTAATGACTCCAAACTACGTAAAGAATATAATTGAAAAGTACTTTATTCAGAATAACCATAAGTTAGTTATGACTATGCTACCAGACCCAAAATTTGATGATGTCTTCAATGAAGCAGAATCTAAACTTCTGAAATCAAAAGTTTCGTCCTTAAGTGCTAACGATAAAGAAGCTATTTATAAAGAAGGCTTAGAGCTTTCCAAAGTTCAGAAGGAGGTGCAGAATCTTGATGTTTTACCATGTTTGAAAATGGAAGAGATCGTAGCGAACAAAACCGCCCCATCATTAAAACATTCTGTATCAGAAACCATACCACTGCAACTTTGTGAGGCGAATACAAATGGCGTAACATATTTCAAAGGAATCTTAGGCACCGACAGCTTGGGCGACAGTGAAAGAAAAATGCTTCCCTTTTTTAATTATATTTTAGATAAATTTGACACTAAGTCGTATAACTACCGTGATTTTGATAAGCATGTCAGCAAATGCACCTCTGGCCTGAGTGCAATTACACACATCACTGAGCATGTCGGGCAATCTGGTCAATATGAGCAGGGTATATTGATCAACAGTCATTGTTTGGACCACAATCTACCTAAAATGCTTGATATATGGCAGGAGATATTTAAAAAACCAAATTTTAGCAACAGCGAAAGAATGGCAATGCTTCTAAATAATTACTGTTCTTCGTTGACAAATGGGATAGTAAGCAGCGGTCACACTTATGCTGTACAGGCGGCCAGGTCTTTAATATCTTCTGTAGATGAATGCAAAGAGAACCTGCTGGGTATCCAACACGTCACGATCATGCAGGAAGTACAAAAAACACAGAAAACTGAAGATATACAGTCGGTAATAGAGAGCATATCAGAAAACGTACTTAAAGGGAATAATTTGCGAGCAGCATTCCATTATTGCAATACAAACAACGATGTCCACGAATACATCGATAAATTCTGCAAGGACCTGTGTAACGCTAATGATAATCAAGAAGTCAATCGCATTAATTGGACAGACTCTAAAAGTATGAACAAGGACAACCGCGGTATACATATAGCCATGAATATACCAGTGAATTTTTGCGCCAAAGTCATACCGACTGTTGCGTACACCCATCCGGATTACGCGAAGCTACGCGTGCTGTCAGTGTTCTTATCGTCGAAATACTTGCACCCGATCGTGCGAGAGCAGAACGGTGCTTACGGTGGCCGCGCCATGCTGTCCTTTGACGGAATCTTCAGTTACTATTCATACAGAGACCCAAATTCTAAAGTCACTTTGGACGTGTTCGACGAAACCGCCAATTGGTTGACAAAAAATGCGAATTTGATTGACGAACAAAACCTCTTCGAGTCCAAGCTAACTATAATGCAGCAAATGGACCAACCGATCGCGGAATACATGAAAGGAGTCGATCTGTTCCTGTACGGGCTGTCTTATGACCTTTGGAAGACGCAAAGGGAAAGAGTTTTGGCCGTGAAAAGCGAGGACTTGGTGGAAGTATGCTTGAAATATCTGACGAACGATAAATGGGCCGCTAAATGTGTAATCGGAGATGGTGGTGCACAGGATTTGAAGAAAGGGGATGAGATGTCTTTGCCTGAATCGTTTCAAGAGGACAGGCTATACTTCGAAGAATGCCTATCATACTTTCAAGATTATCAAAATCTATACAAACACCCGAACACAGAAATATTAGTTGAAAATGTTTTGCAACATATCATTTTGGGAGATTTATCTAAACACGATGTTTTAAATAATAATTTCGAGCTGAATTCAATCGAGGATGAAGTTATGATGAAAACATTTTTGGAGCGCACTCGGAGATTGACAGTGTGTCACGATGACGCTGTCAAAATTGGCGCGGAATTCGATCTCGACGCCCCAGTTGGCATTAAGAAAAAGTATGAAATTGTAAACTTAGCTAAAGAGATCAAAGGAATCTGTGAAGATGTAAACTGTGGGATGGTTGTGGATTTCGGGTCGGGTTTGGGCTATCTGGATCAATATTTACATTCTATTAGTCAAGTCAAAATCTTAGGTTTGGAATGTAACGACAGTCATTATGTCACCGCCAAAAAACGCCAAACGAAATACCACGAGAGTTCGTCAGAGCATATTGGCAGCACAGCCAACGAGTTGTGGAGATAA
Protein
MYSRLRNGFRQTVKHRLASQRSYSNEVLRKKENLSKNLQVGKTIHGFLCTEIESIQEYNMTAYFLTHERTKTEYLHLERDDSNNVFSVGFRTTPLDSMGTPHILEHTVLCGSEKYPVRDPFFKMLNRSLATFMNALTGPDYTFYPFSSQNEIDYRNLQKVYLDAVFRPNLTRLDFLQEGWRLEHSNLEDKNSDLIFKGVVYNEMKGAFSETSSLFGQKFINTILPKGTYGYVSGGDPLCIPQLTHEHLKNFHATYYHPSNSRMYSYGSFPLEANLKFINEAYLSKYEYLDPKNTVVASQERWKEPRRAEMNCRVDQYGGPVEKQNQIAIGYVMSDITNIYETFTMLALAELMIIGPNSAFYKSLIEKNISGGYNSLTGYENQIRDTMFVVGLRDVETSKFETIEKIANQTLEEIYEKGFEKAHIESVLHGFELSIKHQSPKFGLNLLFNLMPLWNHNGPILNALKVNKLLGRLKENLMTPNYVKNIIEKYFIQNNHKLVMTMLPDPKFDDVFNEAESKLLKSKVSSLSANDKEAIYKEGLELSKVQKEVQNLDVLPCLKMEEIVANKTAPSLKHSVSETIPLQLCEANTNGVTYFKGILGTDSLGDSERKMLPFFNYILDKFDTKSYNYRDFDKHVSKCTSGLSAITHITEHVGQSGQYEQGILINSHCLDHNLPKMLDIWQEIFKKPNFSNSERMAMLLNNYCSSLTNGIVSSGHTYAVQAARSLISSVDECKENLLGIQHVTIMQEVQKTQKTEDIQSVIESISENVLKGNNLRAAFHYCNTNNDVHEYIDKFCKDLCNANDNQEVNRINWTDSKSMNKDNRGIHIAMNIPVNFCAKVIPTVAYTHPDYAKLRVLSVFLSSKYLHPIVREQNGAYGGRAMLSFDGIFSYYSYRDPNSKVTLDVFDETANWLTKNANLIDEQNLFESKLTIMQQMDQPIAEYMKGVDLFLYGLSYDLWKTQRERVLAVKSEDLVEVCLKYLTNDKWAAKCVIGDGGAQDLKKGDEMSLPESFQEDRLYFEECLSYFQDYQNLYKHPNTEILVENVLQHIILGDLSKHDVLNNNFELNSIEDEVMMKTFLERTRRLTVCHDDAVKIGAEFDLDAPVGIKKKYEIVNLAKEIKGICEDVNCGMVVDFGSGLGYLDQYLHSISQVKILGLECNDSHYVTAKKRQTKYHESSSEHIGSTANELWR
Summary
Description
ATP-independent protease that degrades mitochondrial transit peptides after their cleavage. Also degrades other unstructured peptides (By similarity).
Cofactor
Zn(2+)
Subunit
Homodimer.
Similarity
Belongs to the peptidase M16 family. PreP subfamily.
Keywords
Complete proteome
Hydrolase
Metal-binding
Metalloprotease
Mitochondrion
Protease
Reference proteome
Transit peptide
Zinc
Feature
chain Presequence protease, mitochondrial
Uniprot
A0A0L7LHC8
A0A2H1W6R8
A0A2W1C1V2
A0A212FAE2
A0A0N1ICM9
A0A194PVB3
+ More
A0A182H7Q2 Q16MK3 A0A1S4FVU1 A0A1Q3FC10 A0A336KNG4 A0A336M954 A0A084WGK0 W5J6N1 A0A182IUV4 B0WCZ9 A0A182PMV6 A0A182FGF3 A0A2M4BBU2 A0A2M4A8U4 A0A2M4BBT7 A0A182LKC8 A0A182R103 Q7Q564 A0A182M5B6 A0A1B0CN86 A0A182R966 A0A182JU28 A0A182UZL9 A0A182U9C1 A0A182I4J6 A0A182X4J9 A0A182NI89 A0A182WBP9 A0A182Y2J1 A0A1B0GC92 A0A2M4BCL2 D6WT63 Q172U8 A0A1I8NAK7 A0A0L0C336 T1PJS7 A0A1I8NNH5 N6U0K3 A0A1B0B5B8 A0A1A9XQR5 A0A1B0A3P2 A0A1A9V231 A0A1A9X4R4 W8BEB1 A0A034WNK7 B4MIZ8 A0A0K8TWX8 A0A0A1WEQ7 A0A0J7L2C7 A0A0B4LES3 Q9V9E3 A0A1W4W039 A0A0J9R7C3 B4R4G3 A0A0J9R6F2 B4ISV3 E2AUJ1 B4IM47 B4KLS3 A0A0N0BKM9 B4J6H6 B4LPH0 B3N434 A0A2A3E120 Q293H1 A0A088ADA3 A0A310S9G4 A0A1W4XDQ5 B3MH58 A0A1Y1MI43 A0A1Y1MLI4 A0A3B0JTF4 A0A232ELQ3 A0A195BPX3 A0A158P3D7 B4GBI4 K7IQA1 N6T3D7 A0A151IIU2 A0A0M5J9W0 E2C3H2 E9IPF6 A0A195EZB5 A0A195DNC7 A0A3L8DTH1 A0A151WYL1 A0A1B0DLU7 A0A3B0J3S4 F4X5G1 A0A1B6DCM2 A0A162SU01 A0A154PJU4 A0A2P8YT83
A0A182H7Q2 Q16MK3 A0A1S4FVU1 A0A1Q3FC10 A0A336KNG4 A0A336M954 A0A084WGK0 W5J6N1 A0A182IUV4 B0WCZ9 A0A182PMV6 A0A182FGF3 A0A2M4BBU2 A0A2M4A8U4 A0A2M4BBT7 A0A182LKC8 A0A182R103 Q7Q564 A0A182M5B6 A0A1B0CN86 A0A182R966 A0A182JU28 A0A182UZL9 A0A182U9C1 A0A182I4J6 A0A182X4J9 A0A182NI89 A0A182WBP9 A0A182Y2J1 A0A1B0GC92 A0A2M4BCL2 D6WT63 Q172U8 A0A1I8NAK7 A0A0L0C336 T1PJS7 A0A1I8NNH5 N6U0K3 A0A1B0B5B8 A0A1A9XQR5 A0A1B0A3P2 A0A1A9V231 A0A1A9X4R4 W8BEB1 A0A034WNK7 B4MIZ8 A0A0K8TWX8 A0A0A1WEQ7 A0A0J7L2C7 A0A0B4LES3 Q9V9E3 A0A1W4W039 A0A0J9R7C3 B4R4G3 A0A0J9R6F2 B4ISV3 E2AUJ1 B4IM47 B4KLS3 A0A0N0BKM9 B4J6H6 B4LPH0 B3N434 A0A2A3E120 Q293H1 A0A088ADA3 A0A310S9G4 A0A1W4XDQ5 B3MH58 A0A1Y1MI43 A0A1Y1MLI4 A0A3B0JTF4 A0A232ELQ3 A0A195BPX3 A0A158P3D7 B4GBI4 K7IQA1 N6T3D7 A0A151IIU2 A0A0M5J9W0 E2C3H2 E9IPF6 A0A195EZB5 A0A195DNC7 A0A3L8DTH1 A0A151WYL1 A0A1B0DLU7 A0A3B0J3S4 F4X5G1 A0A1B6DCM2 A0A162SU01 A0A154PJU4 A0A2P8YT83
EC Number
3.4.24.-
Pubmed
26227816
28756777
22118469
26354079
26483478
17510324
+ More
24438588 20920257 23761445 20966253 12364791 14747013 17210077 25244985 18362917 19820115 25315136 26108605 23537049 24495485 25348373 17994087 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 22936249 17550304 20798317 18057021 15632085 23185243 28004739 28648823 21347285 20075255 21282665 30249741 21719571 29403074
24438588 20920257 23761445 20966253 12364791 14747013 17210077 25244985 18362917 19820115 25315136 26108605 23537049 24495485 25348373 17994087 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 22936249 17550304 20798317 18057021 15632085 23185243 28004739 28648823 21347285 20075255 21282665 30249741 21719571 29403074
EMBL
JTDY01001068
KOB74973.1
ODYU01006703
SOQ48779.1
KZ149912
PZC78083.1
+ More
AGBW02009496 OWR50689.1 KQ460940 KPJ10608.1 KQ459593 KPI96699.1 JXUM01000444 CH477859 EAT35563.1 GFDL01009955 JAV25090.1 UFQS01000716 UFQT01000716 SSX06376.1 SSX26730.1 UFQS01000742 UFQT01000742 SSX06513.1 SSX26862.1 ATLV01023587 KE525344 KFB49344.1 ADMH02002104 ETN59058.1 DS231892 EDS44074.1 GGFJ01001374 MBW50515.1 GGFK01003864 MBW37185.1 GGFJ01001373 MBW50514.1 AXCN02000877 AAAB01008960 EAA11299.4 AXCM01001383 AJWK01019897 APCN01002316 CCAG010007832 GGFJ01001644 MBW50785.1 KQ971354 EFA07169.2 CH477430 EAT41081.1 JRES01000960 KNC26740.1 KA648208 AFP62837.1 APGK01054189 APGK01054190 KB741247 ENN72062.1 JXJN01008671 GAMC01006925 JAB99630.1 GAKP01001801 JAC57151.1 CH963719 EDW72087.1 GDHF01033724 GDHF01029931 JAI18590.1 JAI22383.1 GBXI01017342 JAC96949.1 LBMM01001281 KMQ96589.1 AE013599 AHN55937.1 AY102681 CM002911 KMY91594.1 CM000366 EDX17053.1 KMY91592.1 KMY91591.1 CH891608 EDW99535.2 GL442829 EFN62885.1 CH480926 EDW44094.1 CH933808 EDW09733.1 KRG04872.1 KQ435698 KOX80748.1 CH916367 EDW01977.1 CH940648 EDW61229.1 KRF79860.1 CH954177 EDV59928.2 KZ288467 PBC25398.1 CM000071 EAL24640.3 KRT01424.1 KQ762903 OAD55290.1 CH902619 EDV35817.1 KPU75842.1 GEZM01030775 JAV85333.1 GEZM01030773 JAV85335.1 OUUW01000001 SPP75971.1 NNAY01003517 OXU19286.1 KQ976432 KYM87542.1 ADTU01007654 CH479181 EDW31279.1 ENN72063.1 KQ977459 KYN02578.1 CP012524 ALC40681.1 GL452320 EFN77510.1 GL764503 EFZ17552.1 KQ981905 KYN33471.1 KQ980713 KYN14373.1 QOIP01000005 RLU23058.1 KQ982649 KYQ52990.1 AJVK01016557 AJVK01016558 AJVK01016559 SPP75985.1 GL888716 EGI58317.1 GEDC01013857 JAS23441.1 LRGB01000024 KZS21624.1 KQ434924 KZC11558.1 PYGN01000374 PSN47444.1
AGBW02009496 OWR50689.1 KQ460940 KPJ10608.1 KQ459593 KPI96699.1 JXUM01000444 CH477859 EAT35563.1 GFDL01009955 JAV25090.1 UFQS01000716 UFQT01000716 SSX06376.1 SSX26730.1 UFQS01000742 UFQT01000742 SSX06513.1 SSX26862.1 ATLV01023587 KE525344 KFB49344.1 ADMH02002104 ETN59058.1 DS231892 EDS44074.1 GGFJ01001374 MBW50515.1 GGFK01003864 MBW37185.1 GGFJ01001373 MBW50514.1 AXCN02000877 AAAB01008960 EAA11299.4 AXCM01001383 AJWK01019897 APCN01002316 CCAG010007832 GGFJ01001644 MBW50785.1 KQ971354 EFA07169.2 CH477430 EAT41081.1 JRES01000960 KNC26740.1 KA648208 AFP62837.1 APGK01054189 APGK01054190 KB741247 ENN72062.1 JXJN01008671 GAMC01006925 JAB99630.1 GAKP01001801 JAC57151.1 CH963719 EDW72087.1 GDHF01033724 GDHF01029931 JAI18590.1 JAI22383.1 GBXI01017342 JAC96949.1 LBMM01001281 KMQ96589.1 AE013599 AHN55937.1 AY102681 CM002911 KMY91594.1 CM000366 EDX17053.1 KMY91592.1 KMY91591.1 CH891608 EDW99535.2 GL442829 EFN62885.1 CH480926 EDW44094.1 CH933808 EDW09733.1 KRG04872.1 KQ435698 KOX80748.1 CH916367 EDW01977.1 CH940648 EDW61229.1 KRF79860.1 CH954177 EDV59928.2 KZ288467 PBC25398.1 CM000071 EAL24640.3 KRT01424.1 KQ762903 OAD55290.1 CH902619 EDV35817.1 KPU75842.1 GEZM01030775 JAV85333.1 GEZM01030773 JAV85335.1 OUUW01000001 SPP75971.1 NNAY01003517 OXU19286.1 KQ976432 KYM87542.1 ADTU01007654 CH479181 EDW31279.1 ENN72063.1 KQ977459 KYN02578.1 CP012524 ALC40681.1 GL452320 EFN77510.1 GL764503 EFZ17552.1 KQ981905 KYN33471.1 KQ980713 KYN14373.1 QOIP01000005 RLU23058.1 KQ982649 KYQ52990.1 AJVK01016557 AJVK01016558 AJVK01016559 SPP75985.1 GL888716 EGI58317.1 GEDC01013857 JAS23441.1 LRGB01000024 KZS21624.1 KQ434924 KZC11558.1 PYGN01000374 PSN47444.1
Proteomes
UP000037510
UP000007151
UP000053240
UP000053268
UP000069940
UP000008820
+ More
UP000030765 UP000000673 UP000075880 UP000002320 UP000075885 UP000069272 UP000075882 UP000075886 UP000007062 UP000075883 UP000092461 UP000075900 UP000075881 UP000075903 UP000075902 UP000075840 UP000076407 UP000075884 UP000075920 UP000076408 UP000092444 UP000007266 UP000095301 UP000037069 UP000095300 UP000019118 UP000092460 UP000092443 UP000092445 UP000078200 UP000091820 UP000007798 UP000036403 UP000000803 UP000192221 UP000000304 UP000002282 UP000000311 UP000001292 UP000009192 UP000053105 UP000001070 UP000008792 UP000008711 UP000242457 UP000001819 UP000005203 UP000192223 UP000007801 UP000268350 UP000215335 UP000078540 UP000005205 UP000008744 UP000002358 UP000078542 UP000092553 UP000008237 UP000078541 UP000078492 UP000279307 UP000075809 UP000092462 UP000007755 UP000076858 UP000076502 UP000245037
UP000030765 UP000000673 UP000075880 UP000002320 UP000075885 UP000069272 UP000075882 UP000075886 UP000007062 UP000075883 UP000092461 UP000075900 UP000075881 UP000075903 UP000075902 UP000075840 UP000076407 UP000075884 UP000075920 UP000076408 UP000092444 UP000007266 UP000095301 UP000037069 UP000095300 UP000019118 UP000092460 UP000092443 UP000092445 UP000078200 UP000091820 UP000007798 UP000036403 UP000000803 UP000192221 UP000000304 UP000002282 UP000000311 UP000001292 UP000009192 UP000053105 UP000001070 UP000008792 UP000008711 UP000242457 UP000001819 UP000005203 UP000192223 UP000007801 UP000268350 UP000215335 UP000078540 UP000005205 UP000008744 UP000002358 UP000078542 UP000092553 UP000008237 UP000078541 UP000078492 UP000279307 UP000075809 UP000092462 UP000007755 UP000076858 UP000076502 UP000245037
PRIDE
Pfam
Interpro
IPR011249
Metalloenz_LuxS/M16
+ More
IPR025714 Methyltranfer_dom
IPR007863 Peptidase_M16_C
IPR013578 Peptidase_M16C_assoc
IPR011765 Pept_M16_N
IPR029063 SAM-dependent_MTases
IPR002110 Ankyrin_rpt
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR037191 VPS9_dom_sf
IPR003123 VPS9
IPR001431 Pept_M16_Zn_BS
IPR025714 Methyltranfer_dom
IPR007863 Peptidase_M16_C
IPR013578 Peptidase_M16C_assoc
IPR011765 Pept_M16_N
IPR029063 SAM-dependent_MTases
IPR002110 Ankyrin_rpt
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR037191 VPS9_dom_sf
IPR003123 VPS9
IPR001431 Pept_M16_Zn_BS
Gene 3D
CDD
ProteinModelPortal
A0A0L7LHC8
A0A2H1W6R8
A0A2W1C1V2
A0A212FAE2
A0A0N1ICM9
A0A194PVB3
+ More
A0A182H7Q2 Q16MK3 A0A1S4FVU1 A0A1Q3FC10 A0A336KNG4 A0A336M954 A0A084WGK0 W5J6N1 A0A182IUV4 B0WCZ9 A0A182PMV6 A0A182FGF3 A0A2M4BBU2 A0A2M4A8U4 A0A2M4BBT7 A0A182LKC8 A0A182R103 Q7Q564 A0A182M5B6 A0A1B0CN86 A0A182R966 A0A182JU28 A0A182UZL9 A0A182U9C1 A0A182I4J6 A0A182X4J9 A0A182NI89 A0A182WBP9 A0A182Y2J1 A0A1B0GC92 A0A2M4BCL2 D6WT63 Q172U8 A0A1I8NAK7 A0A0L0C336 T1PJS7 A0A1I8NNH5 N6U0K3 A0A1B0B5B8 A0A1A9XQR5 A0A1B0A3P2 A0A1A9V231 A0A1A9X4R4 W8BEB1 A0A034WNK7 B4MIZ8 A0A0K8TWX8 A0A0A1WEQ7 A0A0J7L2C7 A0A0B4LES3 Q9V9E3 A0A1W4W039 A0A0J9R7C3 B4R4G3 A0A0J9R6F2 B4ISV3 E2AUJ1 B4IM47 B4KLS3 A0A0N0BKM9 B4J6H6 B4LPH0 B3N434 A0A2A3E120 Q293H1 A0A088ADA3 A0A310S9G4 A0A1W4XDQ5 B3MH58 A0A1Y1MI43 A0A1Y1MLI4 A0A3B0JTF4 A0A232ELQ3 A0A195BPX3 A0A158P3D7 B4GBI4 K7IQA1 N6T3D7 A0A151IIU2 A0A0M5J9W0 E2C3H2 E9IPF6 A0A195EZB5 A0A195DNC7 A0A3L8DTH1 A0A151WYL1 A0A1B0DLU7 A0A3B0J3S4 F4X5G1 A0A1B6DCM2 A0A162SU01 A0A154PJU4 A0A2P8YT83
A0A182H7Q2 Q16MK3 A0A1S4FVU1 A0A1Q3FC10 A0A336KNG4 A0A336M954 A0A084WGK0 W5J6N1 A0A182IUV4 B0WCZ9 A0A182PMV6 A0A182FGF3 A0A2M4BBU2 A0A2M4A8U4 A0A2M4BBT7 A0A182LKC8 A0A182R103 Q7Q564 A0A182M5B6 A0A1B0CN86 A0A182R966 A0A182JU28 A0A182UZL9 A0A182U9C1 A0A182I4J6 A0A182X4J9 A0A182NI89 A0A182WBP9 A0A182Y2J1 A0A1B0GC92 A0A2M4BCL2 D6WT63 Q172U8 A0A1I8NAK7 A0A0L0C336 T1PJS7 A0A1I8NNH5 N6U0K3 A0A1B0B5B8 A0A1A9XQR5 A0A1B0A3P2 A0A1A9V231 A0A1A9X4R4 W8BEB1 A0A034WNK7 B4MIZ8 A0A0K8TWX8 A0A0A1WEQ7 A0A0J7L2C7 A0A0B4LES3 Q9V9E3 A0A1W4W039 A0A0J9R7C3 B4R4G3 A0A0J9R6F2 B4ISV3 E2AUJ1 B4IM47 B4KLS3 A0A0N0BKM9 B4J6H6 B4LPH0 B3N434 A0A2A3E120 Q293H1 A0A088ADA3 A0A310S9G4 A0A1W4XDQ5 B3MH58 A0A1Y1MI43 A0A1Y1MLI4 A0A3B0JTF4 A0A232ELQ3 A0A195BPX3 A0A158P3D7 B4GBI4 K7IQA1 N6T3D7 A0A151IIU2 A0A0M5J9W0 E2C3H2 E9IPF6 A0A195EZB5 A0A195DNC7 A0A3L8DTH1 A0A151WYL1 A0A1B0DLU7 A0A3B0J3S4 F4X5G1 A0A1B6DCM2 A0A162SU01 A0A154PJU4 A0A2P8YT83
PDB
4RPU
E-value=0,
Score=1887
Ontologies
GO
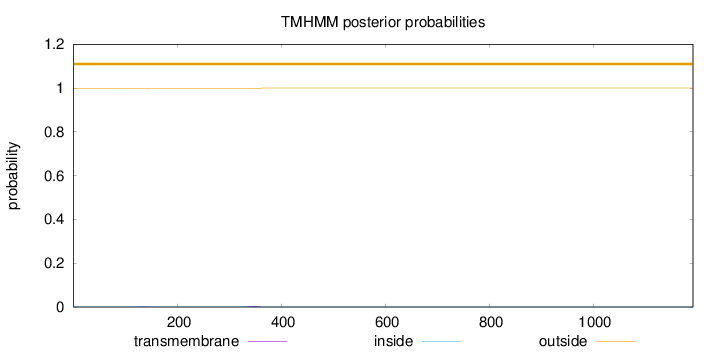
Topology
Subcellular location
Mitochondrion
Length:
1192
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0865400000000001
Exp number, first 60 AAs:
0.0005
Total prob of N-in:
0.00375
outside
1 - 1192
Population Genetic Test Statistics
Pi
256.075575
Theta
183.619253
Tajima's D
0.938659
CLR
0.047539
CSRT
0.64166791660417
Interpretation
Uncertain
