Gene
KWMTBOMO00685
Pre Gene Modal
BGIBMGA003220
Annotation
PREDICTED:_glutamate_decarboxylase_[Bombyx_mori]
Full name
Glutamate decarboxylase
Location in the cell
Cytoplasmic Reliability : 2.694
Sequence
CDS
ATGAAACTTTTTCAGGGTATCCTTCTGAGCTGCAACGCTATGTCGGCTGAATACTTGTTCATGACAGACAAGATCTATGATCCTCGATTCGACACCGGCGACAAGGTGATCCAGTGTGGGAGGCACAATGATATCTTCAAGCTGTGGCTGCAATGGCGCGGAAAGGGTACGTCCGGATTTGAACGCCTCATGGATCGTCTCATGGAGCTATCAGAGTATATGGTGCGCCGCATCAAGGAGCAGCCGGACAAGTTCTACCTCATTTTGGAGCCGGAGCTGGTCAACGTGTCGTTCTGGTATCTGCCGAAGCAGCTGCGAGGGATTCCTCATGATGCCAACAAGGAGATACGTTTGGGGAAGGTGTGTGCCAAGCTGAAGGGAAGAATGATGCAGTCCGGTACCCTGATGGTGGGCTACCAGCCGGACGACCGCCGCCCCAACTTCTTCAGGAACATAATATCGTCGGCGGCTGTCACGGAAAAGGACGTGGACTTCTTACTCTCAGAGATGGACCGCCTCGGTCAGGACATTGTTGTTGAATAA
Protein
MKLFQGILLSCNAMSAEYLFMTDKIYDPRFDTGDKVIQCGRHNDIFKLWLQWRGKGTSGFERLMDRLMELSEYMVRRIKEQPDKFYLILEPELVNVSFWYLPKQLRGIPHDANKEIRLGKVCAKLKGRMMQSGTLMVGYQPDDRRPNFFRNIISSAAVTEKDVDFLLSEMDRLGQDIVVE
Summary
Catalytic Activity
H(+) + L-glutamate = 4-aminobutanoate + CO2
Cofactor
pyridoxal 5'-phosphate
Subunit
Homodimer.
Similarity
Belongs to the group II decarboxylase family.
Keywords
Complete proteome
Decarboxylase
Lyase
Neurotransmitter biosynthesis
Pyridoxal phosphate
Reference proteome
Feature
chain Glutamate decarboxylase
Uniprot
H9J133
A0A0G3VI11
A0A212FH07
A0A2W1BZ30
A0A0N0PBF8
A0A194PTE4
+ More
A0A2A4J514 A0A2H1VPL5 U4UGR0 K7XPW9 N6U7J9 A0A182KH65 A0A026W8B3 F4X0P1 A0A3L8DJR5 D6WRJ1 K7IUX9 A0A195F6W9 A0A1Q3FUC4 A0A232EY01 A0A158NCY8 A0A195B879 T1PLK2 A0A151WYC3 A0A182GBD9 A0A151JAL0 A0A195D0A8 B0XFV4 E2ASG5 T1DQE1 A0A2A3EJ83 V9IEL3 A0A0L7R6C2 K7WYY0 E2BY32 Q16NG0 A0A182JCW1 A0A0M8ZR31 A0A084WRT3 A0A182RVZ8 A0A182FCL3 A0A182WHZ5 A0A182MFY9 A0A182PTR5 A0A1B6DW78 W5JFV1 A0A182N846 U5ELT3 A0A1J1JAW4 A0A1I8N673 A0A1I8PBR8 A0A182XMB7 A0A182UYD2 A0A182TN96 A0A182KPX0 Q7PNL7 A0A182SD50 B4IYX3 A0A182Y2Z6 A0A182I252 A0A087ZN21 A0A1I8PBY7 B4N410 W8B868 A0A182QS79 A0A034V440 M9PH99 B4HTZ3 A0A0J9UE83 B3NG30 B4PHP0 P20228 A0A1Y1K1D6 A0A1W4W6H7 B4KWK0 B3M5G6 A0A1W4WYA5 A0A1A9YEK3 A0A1A9V4B6 A0A1B0B7T2 E0VAE3 B4QQ37 A0A0K8WKJ6 A0A3B0JX11 A0A1A9ZRZ0 A0A0K8VBT3 A0A336LXE2 A0A336MQR1 B4LED0 A0A0M4EH78 A0A1A9W4R4 Q2LZW7 A0A067QYW5 A0A1B6ECJ1 A0A2S2P8Z1 A0A2H8TRP5 O44103 O44102 T1HHW1 J9JXN1
A0A2A4J514 A0A2H1VPL5 U4UGR0 K7XPW9 N6U7J9 A0A182KH65 A0A026W8B3 F4X0P1 A0A3L8DJR5 D6WRJ1 K7IUX9 A0A195F6W9 A0A1Q3FUC4 A0A232EY01 A0A158NCY8 A0A195B879 T1PLK2 A0A151WYC3 A0A182GBD9 A0A151JAL0 A0A195D0A8 B0XFV4 E2ASG5 T1DQE1 A0A2A3EJ83 V9IEL3 A0A0L7R6C2 K7WYY0 E2BY32 Q16NG0 A0A182JCW1 A0A0M8ZR31 A0A084WRT3 A0A182RVZ8 A0A182FCL3 A0A182WHZ5 A0A182MFY9 A0A182PTR5 A0A1B6DW78 W5JFV1 A0A182N846 U5ELT3 A0A1J1JAW4 A0A1I8N673 A0A1I8PBR8 A0A182XMB7 A0A182UYD2 A0A182TN96 A0A182KPX0 Q7PNL7 A0A182SD50 B4IYX3 A0A182Y2Z6 A0A182I252 A0A087ZN21 A0A1I8PBY7 B4N410 W8B868 A0A182QS79 A0A034V440 M9PH99 B4HTZ3 A0A0J9UE83 B3NG30 B4PHP0 P20228 A0A1Y1K1D6 A0A1W4W6H7 B4KWK0 B3M5G6 A0A1W4WYA5 A0A1A9YEK3 A0A1A9V4B6 A0A1B0B7T2 E0VAE3 B4QQ37 A0A0K8WKJ6 A0A3B0JX11 A0A1A9ZRZ0 A0A0K8VBT3 A0A336LXE2 A0A336MQR1 B4LED0 A0A0M4EH78 A0A1A9W4R4 Q2LZW7 A0A067QYW5 A0A1B6ECJ1 A0A2S2P8Z1 A0A2H8TRP5 O44103 O44102 T1HHW1 J9JXN1
EC Number
4.1.1.15
Pubmed
19121390
22118469
28756777
26354079
23537049
23220582
+ More
24508170 21719571 30249741 18362917 19820115 20075255 28648823 21347285 26483478 20798317 17510324 24438588 20920257 23761445 25315136 20966253 12364791 14747013 17210077 17994087 25244985 18057021 24495485 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 1689376 12537569 28004739 20566863 15632085 23185243 24845553 9720289
24508170 21719571 30249741 18362917 19820115 20075255 28648823 21347285 26483478 20798317 17510324 24438588 20920257 23761445 25315136 20966253 12364791 14747013 17210077 17994087 25244985 18057021 24495485 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 1689376 12537569 28004739 20566863 15632085 23185243 24845553 9720289
EMBL
BABH01031468
KP657640
AKL78865.1
AGBW02008572
OWR53022.1
KZ149912
+ More
PZC78090.1 KQ460940 KPJ10601.1 KQ459593 KPI96691.1 NWSH01003345 PCG66503.1 ODYU01003671 SOQ42738.1 KB632188 KI208416 KI209198 ERL89786.1 ERL95860.1 ERL95995.1 KC134204 AFX68714.1 APGK01045889 KB741039 ENN74557.1 KK107347 EZA52347.1 GL888498 EGI60033.1 QOIP01000007 RLU20637.1 KQ971351 EFA06442.1 AAZX01001184 KQ981744 KYN36345.1 GFDL01003845 JAV31200.1 NNAY01001694 OXU23203.1 ADTU01012083 ADTU01012084 KQ976558 KYM80711.1 KA648955 AFP63584.1 KQ982650 KYQ52902.1 JXUM01052660 JXUM01052661 KQ561744 KXJ77663.1 KQ979266 KYN22051.1 KQ977012 KYN06353.1 DS232965 EDS27073.1 GL442298 EFN63631.1 GAMD01001210 JAB00381.1 KZ288242 PBC31249.1 JR039580 AEY58764.1 KQ414646 KOC66364.1 KC134205 AFX68715.1 GL451412 EFN79405.1 CH477824 EAT35903.1 KQ435896 KOX69217.1 ATLV01026194 KE525407 KFB52927.1 AXCM01004956 AXCM01004957 AXCM01004958 GEDC01007378 JAS29920.1 ADMH02001620 ETN61760.1 GANO01001281 JAB58590.1 CVRI01000075 CRL08726.1 AAAB01008960 EAA11955.3 CH916366 EDV97681.1 APCN01000120 CH964095 EDW79365.1 KRF99053.1 GAMC01017141 GAMC01017140 GAMC01017139 JAB89416.1 AXCN02000918 GAKP01022035 GAKP01022034 GAKP01022033 JAC36917.1 AE014296 AGB94091.1 CH480817 EDW50414.1 CM002912 KMY97484.1 KMY97485.1 KMY97486.1 CH954178 EDV50859.1 KQS43543.1 KQS43544.1 CM000159 EDW93349.1 KRK01117.1 KRK01118.1 X76198 AY089526 GEZM01097269 JAV54301.1 CH933809 EDW19629.1 KRG06707.1 CH902618 EDV39576.1 KPU78089.1 JXJN01009715 JXJN01009716 DS235005 EEB10349.1 CM000363 EDX09157.1 GDHF01000648 JAI51666.1 OUUW01000009 SPP84962.1 GDHF01017159 GDHF01016022 GDHF01004976 JAI35155.1 JAI36292.1 JAI47338.1 UFQT01000279 SSX22712.1 UFQT01001601 SSX31123.1 CH940647 EDW70106.1 KRF84711.1 CP012525 ALC43026.1 CH379069 EAL31170.1 KRT07941.1 KK853095 KDR11483.1 GEDC01001640 JAS35658.1 GGMR01012757 MBY25376.1 GFXV01004756 MBW16561.1 AF025808 AAB87893.1 AF025807 AAB87892.1 ACPB03000438 ABLF02022308 ABLF02022309
PZC78090.1 KQ460940 KPJ10601.1 KQ459593 KPI96691.1 NWSH01003345 PCG66503.1 ODYU01003671 SOQ42738.1 KB632188 KI208416 KI209198 ERL89786.1 ERL95860.1 ERL95995.1 KC134204 AFX68714.1 APGK01045889 KB741039 ENN74557.1 KK107347 EZA52347.1 GL888498 EGI60033.1 QOIP01000007 RLU20637.1 KQ971351 EFA06442.1 AAZX01001184 KQ981744 KYN36345.1 GFDL01003845 JAV31200.1 NNAY01001694 OXU23203.1 ADTU01012083 ADTU01012084 KQ976558 KYM80711.1 KA648955 AFP63584.1 KQ982650 KYQ52902.1 JXUM01052660 JXUM01052661 KQ561744 KXJ77663.1 KQ979266 KYN22051.1 KQ977012 KYN06353.1 DS232965 EDS27073.1 GL442298 EFN63631.1 GAMD01001210 JAB00381.1 KZ288242 PBC31249.1 JR039580 AEY58764.1 KQ414646 KOC66364.1 KC134205 AFX68715.1 GL451412 EFN79405.1 CH477824 EAT35903.1 KQ435896 KOX69217.1 ATLV01026194 KE525407 KFB52927.1 AXCM01004956 AXCM01004957 AXCM01004958 GEDC01007378 JAS29920.1 ADMH02001620 ETN61760.1 GANO01001281 JAB58590.1 CVRI01000075 CRL08726.1 AAAB01008960 EAA11955.3 CH916366 EDV97681.1 APCN01000120 CH964095 EDW79365.1 KRF99053.1 GAMC01017141 GAMC01017140 GAMC01017139 JAB89416.1 AXCN02000918 GAKP01022035 GAKP01022034 GAKP01022033 JAC36917.1 AE014296 AGB94091.1 CH480817 EDW50414.1 CM002912 KMY97484.1 KMY97485.1 KMY97486.1 CH954178 EDV50859.1 KQS43543.1 KQS43544.1 CM000159 EDW93349.1 KRK01117.1 KRK01118.1 X76198 AY089526 GEZM01097269 JAV54301.1 CH933809 EDW19629.1 KRG06707.1 CH902618 EDV39576.1 KPU78089.1 JXJN01009715 JXJN01009716 DS235005 EEB10349.1 CM000363 EDX09157.1 GDHF01000648 JAI51666.1 OUUW01000009 SPP84962.1 GDHF01017159 GDHF01016022 GDHF01004976 JAI35155.1 JAI36292.1 JAI47338.1 UFQT01000279 SSX22712.1 UFQT01001601 SSX31123.1 CH940647 EDW70106.1 KRF84711.1 CP012525 ALC43026.1 CH379069 EAL31170.1 KRT07941.1 KK853095 KDR11483.1 GEDC01001640 JAS35658.1 GGMR01012757 MBY25376.1 GFXV01004756 MBW16561.1 AF025808 AAB87893.1 AF025807 AAB87892.1 ACPB03000438 ABLF02022308 ABLF02022309
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000030742
+ More
UP000019118 UP000075881 UP000053097 UP000007755 UP000279307 UP000007266 UP000002358 UP000078541 UP000215335 UP000005205 UP000078540 UP000075809 UP000069940 UP000249989 UP000078492 UP000078542 UP000002320 UP000000311 UP000242457 UP000053825 UP000008237 UP000008820 UP000075880 UP000053105 UP000030765 UP000075900 UP000069272 UP000075920 UP000075883 UP000075885 UP000000673 UP000075884 UP000183832 UP000095301 UP000095300 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075901 UP000001070 UP000076408 UP000075840 UP000005203 UP000007798 UP000075886 UP000000803 UP000001292 UP000008711 UP000002282 UP000192221 UP000009192 UP000007801 UP000192223 UP000092443 UP000078200 UP000092460 UP000009046 UP000000304 UP000268350 UP000092445 UP000008792 UP000092553 UP000091820 UP000001819 UP000027135 UP000015103 UP000007819
UP000019118 UP000075881 UP000053097 UP000007755 UP000279307 UP000007266 UP000002358 UP000078541 UP000215335 UP000005205 UP000078540 UP000075809 UP000069940 UP000249989 UP000078492 UP000078542 UP000002320 UP000000311 UP000242457 UP000053825 UP000008237 UP000008820 UP000075880 UP000053105 UP000030765 UP000075900 UP000069272 UP000075920 UP000075883 UP000075885 UP000000673 UP000075884 UP000183832 UP000095301 UP000095300 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075901 UP000001070 UP000076408 UP000075840 UP000005203 UP000007798 UP000075886 UP000000803 UP000001292 UP000008711 UP000002282 UP000192221 UP000009192 UP000007801 UP000192223 UP000092443 UP000078200 UP000092460 UP000009046 UP000000304 UP000268350 UP000092445 UP000008792 UP000092553 UP000091820 UP000001819 UP000027135 UP000015103 UP000007819
Pfam
PF00282 Pyridoxal_deC
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
H9J133
A0A0G3VI11
A0A212FH07
A0A2W1BZ30
A0A0N0PBF8
A0A194PTE4
+ More
A0A2A4J514 A0A2H1VPL5 U4UGR0 K7XPW9 N6U7J9 A0A182KH65 A0A026W8B3 F4X0P1 A0A3L8DJR5 D6WRJ1 K7IUX9 A0A195F6W9 A0A1Q3FUC4 A0A232EY01 A0A158NCY8 A0A195B879 T1PLK2 A0A151WYC3 A0A182GBD9 A0A151JAL0 A0A195D0A8 B0XFV4 E2ASG5 T1DQE1 A0A2A3EJ83 V9IEL3 A0A0L7R6C2 K7WYY0 E2BY32 Q16NG0 A0A182JCW1 A0A0M8ZR31 A0A084WRT3 A0A182RVZ8 A0A182FCL3 A0A182WHZ5 A0A182MFY9 A0A182PTR5 A0A1B6DW78 W5JFV1 A0A182N846 U5ELT3 A0A1J1JAW4 A0A1I8N673 A0A1I8PBR8 A0A182XMB7 A0A182UYD2 A0A182TN96 A0A182KPX0 Q7PNL7 A0A182SD50 B4IYX3 A0A182Y2Z6 A0A182I252 A0A087ZN21 A0A1I8PBY7 B4N410 W8B868 A0A182QS79 A0A034V440 M9PH99 B4HTZ3 A0A0J9UE83 B3NG30 B4PHP0 P20228 A0A1Y1K1D6 A0A1W4W6H7 B4KWK0 B3M5G6 A0A1W4WYA5 A0A1A9YEK3 A0A1A9V4B6 A0A1B0B7T2 E0VAE3 B4QQ37 A0A0K8WKJ6 A0A3B0JX11 A0A1A9ZRZ0 A0A0K8VBT3 A0A336LXE2 A0A336MQR1 B4LED0 A0A0M4EH78 A0A1A9W4R4 Q2LZW7 A0A067QYW5 A0A1B6ECJ1 A0A2S2P8Z1 A0A2H8TRP5 O44103 O44102 T1HHW1 J9JXN1
A0A2A4J514 A0A2H1VPL5 U4UGR0 K7XPW9 N6U7J9 A0A182KH65 A0A026W8B3 F4X0P1 A0A3L8DJR5 D6WRJ1 K7IUX9 A0A195F6W9 A0A1Q3FUC4 A0A232EY01 A0A158NCY8 A0A195B879 T1PLK2 A0A151WYC3 A0A182GBD9 A0A151JAL0 A0A195D0A8 B0XFV4 E2ASG5 T1DQE1 A0A2A3EJ83 V9IEL3 A0A0L7R6C2 K7WYY0 E2BY32 Q16NG0 A0A182JCW1 A0A0M8ZR31 A0A084WRT3 A0A182RVZ8 A0A182FCL3 A0A182WHZ5 A0A182MFY9 A0A182PTR5 A0A1B6DW78 W5JFV1 A0A182N846 U5ELT3 A0A1J1JAW4 A0A1I8N673 A0A1I8PBR8 A0A182XMB7 A0A182UYD2 A0A182TN96 A0A182KPX0 Q7PNL7 A0A182SD50 B4IYX3 A0A182Y2Z6 A0A182I252 A0A087ZN21 A0A1I8PBY7 B4N410 W8B868 A0A182QS79 A0A034V440 M9PH99 B4HTZ3 A0A0J9UE83 B3NG30 B4PHP0 P20228 A0A1Y1K1D6 A0A1W4W6H7 B4KWK0 B3M5G6 A0A1W4WYA5 A0A1A9YEK3 A0A1A9V4B6 A0A1B0B7T2 E0VAE3 B4QQ37 A0A0K8WKJ6 A0A3B0JX11 A0A1A9ZRZ0 A0A0K8VBT3 A0A336LXE2 A0A336MQR1 B4LED0 A0A0M4EH78 A0A1A9W4R4 Q2LZW7 A0A067QYW5 A0A1B6ECJ1 A0A2S2P8Z1 A0A2H8TRP5 O44103 O44102 T1HHW1 J9JXN1
PDB
3VP6
E-value=4.05138e-53,
Score=521
Ontologies
PATHWAY
00250
Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00430 Taurine and hypotaurine metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00430 Taurine and hypotaurine metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.522714
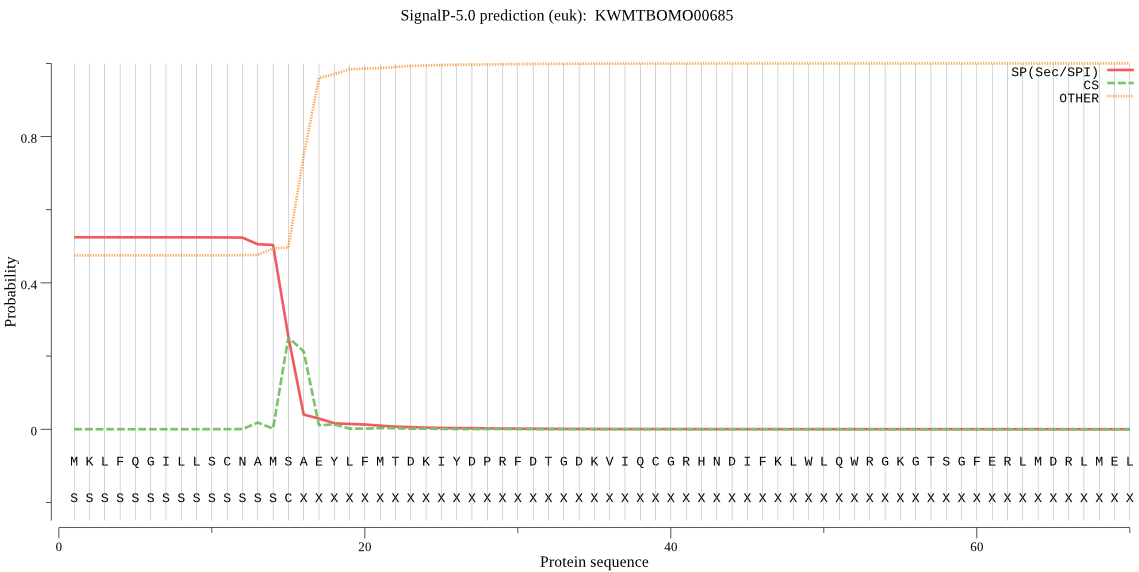
Length:
180
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02543
Exp number, first 60 AAs:
0.02224
Total prob of N-in:
0.01651
outside
1 - 180

Population Genetic Test Statistics
Pi
231.263821
Theta
172.75889
Tajima's D
1.194438
CLR
0
CSRT
0.715164241787911
Interpretation
Uncertain
