Gene
KWMTBOMO00675
Pre Gene Modal
BGIBMGA003286
Annotation
PREDICTED:_neogenin_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.34 PlasmaMembrane Reliability : 1.062
Sequence
CDS
ATGAGCTCGTCACTTGGTGGAGGAGAAGAGGATCCCGGAGTGAGGTTCACGACGGAGCCAAGCGACGCAGTGGTCCTTCCCGGTGAAGATGCGTTGCTTCTCTGCGCGGCTGCTTCCAAGCTGCCCGTGCGTCTGGCGTGGCGGTACAGCGCCAGTTCCCTGCCCACCAGAGACCACGGCGTCTCTATTAGCGATAAGTCCAGGAAGCAACTCCTCAACGGCTCCCTTCTGATAGAGAGCATGACAGCGTCCTTGGCGGGCCAGTACCAGTGTGTGGCCACCGTGGACGGTATCGGTACCATCGTCTCAAGAGCCGCCACCGTGTTCTTAGCTGAGGTGCCAGTGTTGTCGGGCACCCCGCGCAGCGTGCTGTCCGCGGTGGGGGGCGTGGCGCTGCTGCCGTGCGGGATACAGGCCCCGCCGAGACTGGCCCTCAGAGTCGTGCCCGCCGCCAACACCGACAGGAGGGCGTACGGCGCCGCCAAGCTACTCAACACTCCGCCCCTCCTCAAACTCAATGTGACCTGGTTAAAGAACGGCTCCCCGGTCCGCACGGAGAGCGCCCGCGTCTCCATCACGCCCAGCGGAGCCTTGGAGATAGAGCCGCTGCGGCACCACGACGCCGCCTCCTACAGATGTCGTGTTTCTATCACACCTCATCTATATCGGGTAAGCGATGAGATAGAGCTACGCGTGAGTGCTGACACTCCCAACACGGAGGCGGCCCCCAGGTTCGTCGCGATGCCGCAATCCCTCACTGTCATGGAAGGCGGCTCCGCGACCCTGGACTGCGCCGCCATCGGGAACCCTAAACCAGAGATCATTTGGCTCAACAACGGAGTCGCTATCGACTTAAGCGACCTGGACTCGCGGCTGTACCTGGTGGGGTCGGGCTCGCTGCGGCTGCGGGCGGCGCGCGCGGGGGACGCCGGCGCCTACACGTGCCGCGCCCACAGCCGCCTCGACTCCAGCGACCACACCGCGCACCTGCACGTGCTGGTCGGTACACACGCGCCCTACATACATAAACTGTCTTATCCATGTTTATCCGTCTTTGCACCTTAA
Protein
MSSSLGGGEEDPGVRFTTEPSDAVVLPGEDALLLCAAASKLPVRLAWRYSASSLPTRDHGVSISDKSRKQLLNGSLLIESMTASLAGQYQCVATVDGIGTIVSRAATVFLAEVPVLSGTPRSVLSAVGGVALLPCGIQAPPRLALRVVPAANTDRRAYGAAKLLNTPPLLKLNVTWLKNGSPVRTESARVSITPSGALEIEPLRHHDAASYRCRVSITPHLYRVSDEIELRVSADTPNTEAAPRFVAMPQSLTVMEGGSATLDCAAIGNPKPEIIWLNNGVAIDLSDLDSRLYLVGSGSLRLRAARAGDAGAYTCRAHSRLDSSDHTAHLHVLVGTHAPYIHKLSYPCLSVFAP
Summary
Uniprot
H9J199
A0A2H1WRY9
A0A194PTN9
A0A0N1PFE0
A0A212F3D5
A0A2W1BWU2
+ More
A0A2A4JUT9 A0A2A4JTN1 A0A212F1J6 A0A158P3C4 A0A195CWM6 A0A195BN55 A0A195DFJ3 F4W5X3 E2C3E0 A0A195EYT9 A0A224XGP9 A0A151XH75 A0A182KEB2 A0A182TTA5 A0A3L8DEQ7 A0A232FFG1 K7J3C6 V5I8X2 A0A1W4X9F8 A0A1W4X877 A0A087ZSG0 A0A182Y366 A0A1Y1M7T4 A0A182QE71 T1I1E7 A0A182FI15 A0A154P1U5 A0A146M2A7 A0A1B6DDS9 A0A182P0Y0 W5JMC4 A0A2A3E7S9 Q7Q623 A0A182XM71 A7UU12 A0A182M1U9 A0A0T6BEU8 A0A182R7X4 A0A182NME2 A0A0L7RCW9 A0A1J1HDZ4 A0A182WHR9 A0A310SL89 A0A147BBM2 A0A164QIC7 E2A6Y7 A0A182J115 A0A0P5UX89 A0A0P5KYM7 A0A0P6ES54 A0A0P6GET6 A0A0P5H937 A0A0P5DQP6 A0A0P5KKN1 A0A0P4Y0X2 A0A0P5NYW2 A0A0P5Q443 A0A0P5YH31 A0A0N8ECS1 A0A0P5XFH3 A0A0P5X1H7 A0A0P5UZW4 A0A0P5QT45 A0A0P5ANJ4 A0A0P5XLL3 A0A0N8D858 A0A0P5BPA1 A0A0P5S921 A0A0P5EVV4 A0A0P4WNJ2 A0A0N8B6J0 A0A0P4XSM5 A0A0P5NQE8 A0A0P5PQ66 A0A0P5TTV7 A0A0P6E2W6 A0A0N8CSI5 A0A0P5C0E8 A0A0P5J9C0 A0A0P6INE5 B7QJ27 A0A0P5RFN3 A0A0P5VXG0 A0A084WLA1 A0A0P5WMS4 A0A0P5NAK5 A0A0P5J3L4 A0A0P5J272 A0A0P6EEU7 A0A0P5SFR1 A0A0P5G0C7 A0A0P6IJI1 A0A0P6F6H1 A0A0P5C0B7
A0A2A4JUT9 A0A2A4JTN1 A0A212F1J6 A0A158P3C4 A0A195CWM6 A0A195BN55 A0A195DFJ3 F4W5X3 E2C3E0 A0A195EYT9 A0A224XGP9 A0A151XH75 A0A182KEB2 A0A182TTA5 A0A3L8DEQ7 A0A232FFG1 K7J3C6 V5I8X2 A0A1W4X9F8 A0A1W4X877 A0A087ZSG0 A0A182Y366 A0A1Y1M7T4 A0A182QE71 T1I1E7 A0A182FI15 A0A154P1U5 A0A146M2A7 A0A1B6DDS9 A0A182P0Y0 W5JMC4 A0A2A3E7S9 Q7Q623 A0A182XM71 A7UU12 A0A182M1U9 A0A0T6BEU8 A0A182R7X4 A0A182NME2 A0A0L7RCW9 A0A1J1HDZ4 A0A182WHR9 A0A310SL89 A0A147BBM2 A0A164QIC7 E2A6Y7 A0A182J115 A0A0P5UX89 A0A0P5KYM7 A0A0P6ES54 A0A0P6GET6 A0A0P5H937 A0A0P5DQP6 A0A0P5KKN1 A0A0P4Y0X2 A0A0P5NYW2 A0A0P5Q443 A0A0P5YH31 A0A0N8ECS1 A0A0P5XFH3 A0A0P5X1H7 A0A0P5UZW4 A0A0P5QT45 A0A0P5ANJ4 A0A0P5XLL3 A0A0N8D858 A0A0P5BPA1 A0A0P5S921 A0A0P5EVV4 A0A0P4WNJ2 A0A0N8B6J0 A0A0P4XSM5 A0A0P5NQE8 A0A0P5PQ66 A0A0P5TTV7 A0A0P6E2W6 A0A0N8CSI5 A0A0P5C0E8 A0A0P5J9C0 A0A0P6INE5 B7QJ27 A0A0P5RFN3 A0A0P5VXG0 A0A084WLA1 A0A0P5WMS4 A0A0P5NAK5 A0A0P5J3L4 A0A0P5J272 A0A0P6EEU7 A0A0P5SFR1 A0A0P5G0C7 A0A0P6IJI1 A0A0P6F6H1 A0A0P5C0B7
Pubmed
EMBL
BABH01031504
ODYU01010601
SOQ55821.1
KQ459593
KPI96682.1
KQ460940
+ More
KPJ10593.1 AGBW02010579 OWR48245.1 KZ149912 PZC78094.1 NWSH01000602 PCG75394.1 PCG75395.1 AGBW02010859 OWR47615.1 ADTU01007494 ADTU01007495 KQ977185 KYN05040.1 KQ976432 KYM87460.1 KQ980903 KYN11653.1 GL887695 EGI70449.1 GL452320 EFN77478.1 KQ981905 KYN33388.1 GFTR01008796 JAW07630.1 KQ982130 KYQ59759.1 QOIP01000009 RLU18925.1 NNAY01000313 OXU29293.1 GALX01003940 JAB64526.1 GEZM01041004 JAV80660.1 AXCN02000933 ACPB03008038 ACPB03008039 KQ434796 KZC05827.1 GDHC01005722 JAQ12907.1 GEDC01013469 JAS23829.1 ADMH02000632 ETN65527.1 KZ288340 PBC27758.1 AAAB01008960 EAA11689.4 EDO63788.1 AXCM01004109 AXCM01004110 LJIG01001098 KRT85842.1 KQ414615 KOC68698.1 CVRI01000001 CRK86213.1 KQ764883 OAD54309.1 GEGO01007237 JAR88167.1 LRGB01002391 KZS07779.1 GL437203 EFN70853.1 GDIP01110273 JAL93441.1 GDIQ01185937 JAK65788.1 GDIQ01058639 JAN36098.1 GDIQ01034246 JAN60491.1 GDIQ01229724 JAK22001.1 GDIP01152172 JAJ71230.1 GDIQ01184186 JAK67539.1 GDIP01235312 JAI88089.1 GDIQ01135863 JAL15863.1 GDIQ01140560 JAL11166.1 GDIP01059384 JAM44331.1 GDIQ01039500 JAN55237.1 GDIP01072926 JAM30789.1 GDIP01081175 GDIP01078678 JAM25037.1 GDIP01122916 JAL80798.1 GDIQ01116418 JAL35308.1 GDIP01196360 GDIP01084931 GDIP01083176 JAJ27042.1 GDIP01137270 GDIP01071730 GDIP01070276 JAM33439.1 GDIP01059383 JAM44332.1 GDIP01182210 JAJ41192.1 GDIQ01097020 JAL54706.1 GDIQ01269373 JAJ82351.1 GDIP01253851 GDIP01195310 GDIP01193958 JAI69550.1 GDIQ01207939 JAK43786.1 GDIP01238567 GDIP01235311 JAI84834.1 GDIQ01160748 JAK90977.1 GDIQ01135862 JAL15864.1 GDIP01124076 JAL79638.1 GDIQ01068533 JAN26204.1 GDIP01103167 JAM00548.1 GDIP01177932 JAJ45470.1 GDIQ01205141 JAK46584.1 GDIQ01154702 GDIQ01029003 JAN65734.1 ABJB010044607 ABJB010084407 ABJB010453184 ABJB010719693 ABJB010798131 ABJB011007751 ABJB011066376 DS949082 EEC18849.1 GDIQ01110719 JAL41007.1 GDIP01094724 JAM08991.1 ATLV01024204 KE525350 KFB50995.1 GDIP01084930 JAM18785.1 GDIQ01151801 JAK99924.1 GDIQ01205142 JAK46583.1 GDIQ01207938 JAK43787.1 GDIQ01077928 JAN16809.1 GDIQ01092373 JAL59353.1 GDIQ01269374 JAJ82350.1 GDIQ01005317 JAN89420.1 GDIQ01052690 JAN42047.1 GDIP01177931 JAJ45471.1
KPJ10593.1 AGBW02010579 OWR48245.1 KZ149912 PZC78094.1 NWSH01000602 PCG75394.1 PCG75395.1 AGBW02010859 OWR47615.1 ADTU01007494 ADTU01007495 KQ977185 KYN05040.1 KQ976432 KYM87460.1 KQ980903 KYN11653.1 GL887695 EGI70449.1 GL452320 EFN77478.1 KQ981905 KYN33388.1 GFTR01008796 JAW07630.1 KQ982130 KYQ59759.1 QOIP01000009 RLU18925.1 NNAY01000313 OXU29293.1 GALX01003940 JAB64526.1 GEZM01041004 JAV80660.1 AXCN02000933 ACPB03008038 ACPB03008039 KQ434796 KZC05827.1 GDHC01005722 JAQ12907.1 GEDC01013469 JAS23829.1 ADMH02000632 ETN65527.1 KZ288340 PBC27758.1 AAAB01008960 EAA11689.4 EDO63788.1 AXCM01004109 AXCM01004110 LJIG01001098 KRT85842.1 KQ414615 KOC68698.1 CVRI01000001 CRK86213.1 KQ764883 OAD54309.1 GEGO01007237 JAR88167.1 LRGB01002391 KZS07779.1 GL437203 EFN70853.1 GDIP01110273 JAL93441.1 GDIQ01185937 JAK65788.1 GDIQ01058639 JAN36098.1 GDIQ01034246 JAN60491.1 GDIQ01229724 JAK22001.1 GDIP01152172 JAJ71230.1 GDIQ01184186 JAK67539.1 GDIP01235312 JAI88089.1 GDIQ01135863 JAL15863.1 GDIQ01140560 JAL11166.1 GDIP01059384 JAM44331.1 GDIQ01039500 JAN55237.1 GDIP01072926 JAM30789.1 GDIP01081175 GDIP01078678 JAM25037.1 GDIP01122916 JAL80798.1 GDIQ01116418 JAL35308.1 GDIP01196360 GDIP01084931 GDIP01083176 JAJ27042.1 GDIP01137270 GDIP01071730 GDIP01070276 JAM33439.1 GDIP01059383 JAM44332.1 GDIP01182210 JAJ41192.1 GDIQ01097020 JAL54706.1 GDIQ01269373 JAJ82351.1 GDIP01253851 GDIP01195310 GDIP01193958 JAI69550.1 GDIQ01207939 JAK43786.1 GDIP01238567 GDIP01235311 JAI84834.1 GDIQ01160748 JAK90977.1 GDIQ01135862 JAL15864.1 GDIP01124076 JAL79638.1 GDIQ01068533 JAN26204.1 GDIP01103167 JAM00548.1 GDIP01177932 JAJ45470.1 GDIQ01205141 JAK46584.1 GDIQ01154702 GDIQ01029003 JAN65734.1 ABJB010044607 ABJB010084407 ABJB010453184 ABJB010719693 ABJB010798131 ABJB011007751 ABJB011066376 DS949082 EEC18849.1 GDIQ01110719 JAL41007.1 GDIP01094724 JAM08991.1 ATLV01024204 KE525350 KFB50995.1 GDIP01084930 JAM18785.1 GDIQ01151801 JAK99924.1 GDIQ01205142 JAK46583.1 GDIQ01207938 JAK43787.1 GDIQ01077928 JAN16809.1 GDIQ01092373 JAL59353.1 GDIQ01269374 JAJ82350.1 GDIQ01005317 JAN89420.1 GDIQ01052690 JAN42047.1 GDIP01177931 JAJ45471.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000005205
+ More
UP000078542 UP000078540 UP000078492 UP000007755 UP000008237 UP000078541 UP000075809 UP000075881 UP000075902 UP000279307 UP000215335 UP000002358 UP000192223 UP000005203 UP000076408 UP000075886 UP000015103 UP000069272 UP000076502 UP000075885 UP000000673 UP000242457 UP000007062 UP000076407 UP000075883 UP000075900 UP000075884 UP000053825 UP000183832 UP000075920 UP000076858 UP000000311 UP000075880 UP000001555 UP000030765
UP000078542 UP000078540 UP000078492 UP000007755 UP000008237 UP000078541 UP000075809 UP000075881 UP000075902 UP000279307 UP000215335 UP000002358 UP000192223 UP000005203 UP000076408 UP000075886 UP000015103 UP000069272 UP000076502 UP000075885 UP000000673 UP000242457 UP000007062 UP000076407 UP000075883 UP000075900 UP000075884 UP000053825 UP000183832 UP000075920 UP000076858 UP000000311 UP000075880 UP000001555 UP000030765
PRIDE
Interpro
IPR003598
Ig_sub2
+ More
IPR036179 Ig-like_dom_sf
IPR033024 Neogenin
IPR013098 Ig_I-set
IPR036116 FN3_sf
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR010560 Neogenin_C
IPR013151 Immunoglobulin
IPR003529 Hematopoietin_rcpt_Gp130_CS
IPR013106 Ig_V-set
IPR036396 Cyt_P450_sf
IPR036179 Ig-like_dom_sf
IPR033024 Neogenin
IPR013098 Ig_I-set
IPR036116 FN3_sf
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR010560 Neogenin_C
IPR013151 Immunoglobulin
IPR003529 Hematopoietin_rcpt_Gp130_CS
IPR013106 Ig_V-set
IPR036396 Cyt_P450_sf
Gene 3D
CDD
ProteinModelPortal
H9J199
A0A2H1WRY9
A0A194PTN9
A0A0N1PFE0
A0A212F3D5
A0A2W1BWU2
+ More
A0A2A4JUT9 A0A2A4JTN1 A0A212F1J6 A0A158P3C4 A0A195CWM6 A0A195BN55 A0A195DFJ3 F4W5X3 E2C3E0 A0A195EYT9 A0A224XGP9 A0A151XH75 A0A182KEB2 A0A182TTA5 A0A3L8DEQ7 A0A232FFG1 K7J3C6 V5I8X2 A0A1W4X9F8 A0A1W4X877 A0A087ZSG0 A0A182Y366 A0A1Y1M7T4 A0A182QE71 T1I1E7 A0A182FI15 A0A154P1U5 A0A146M2A7 A0A1B6DDS9 A0A182P0Y0 W5JMC4 A0A2A3E7S9 Q7Q623 A0A182XM71 A7UU12 A0A182M1U9 A0A0T6BEU8 A0A182R7X4 A0A182NME2 A0A0L7RCW9 A0A1J1HDZ4 A0A182WHR9 A0A310SL89 A0A147BBM2 A0A164QIC7 E2A6Y7 A0A182J115 A0A0P5UX89 A0A0P5KYM7 A0A0P6ES54 A0A0P6GET6 A0A0P5H937 A0A0P5DQP6 A0A0P5KKN1 A0A0P4Y0X2 A0A0P5NYW2 A0A0P5Q443 A0A0P5YH31 A0A0N8ECS1 A0A0P5XFH3 A0A0P5X1H7 A0A0P5UZW4 A0A0P5QT45 A0A0P5ANJ4 A0A0P5XLL3 A0A0N8D858 A0A0P5BPA1 A0A0P5S921 A0A0P5EVV4 A0A0P4WNJ2 A0A0N8B6J0 A0A0P4XSM5 A0A0P5NQE8 A0A0P5PQ66 A0A0P5TTV7 A0A0P6E2W6 A0A0N8CSI5 A0A0P5C0E8 A0A0P5J9C0 A0A0P6INE5 B7QJ27 A0A0P5RFN3 A0A0P5VXG0 A0A084WLA1 A0A0P5WMS4 A0A0P5NAK5 A0A0P5J3L4 A0A0P5J272 A0A0P6EEU7 A0A0P5SFR1 A0A0P5G0C7 A0A0P6IJI1 A0A0P6F6H1 A0A0P5C0B7
A0A2A4JUT9 A0A2A4JTN1 A0A212F1J6 A0A158P3C4 A0A195CWM6 A0A195BN55 A0A195DFJ3 F4W5X3 E2C3E0 A0A195EYT9 A0A224XGP9 A0A151XH75 A0A182KEB2 A0A182TTA5 A0A3L8DEQ7 A0A232FFG1 K7J3C6 V5I8X2 A0A1W4X9F8 A0A1W4X877 A0A087ZSG0 A0A182Y366 A0A1Y1M7T4 A0A182QE71 T1I1E7 A0A182FI15 A0A154P1U5 A0A146M2A7 A0A1B6DDS9 A0A182P0Y0 W5JMC4 A0A2A3E7S9 Q7Q623 A0A182XM71 A7UU12 A0A182M1U9 A0A0T6BEU8 A0A182R7X4 A0A182NME2 A0A0L7RCW9 A0A1J1HDZ4 A0A182WHR9 A0A310SL89 A0A147BBM2 A0A164QIC7 E2A6Y7 A0A182J115 A0A0P5UX89 A0A0P5KYM7 A0A0P6ES54 A0A0P6GET6 A0A0P5H937 A0A0P5DQP6 A0A0P5KKN1 A0A0P4Y0X2 A0A0P5NYW2 A0A0P5Q443 A0A0P5YH31 A0A0N8ECS1 A0A0P5XFH3 A0A0P5X1H7 A0A0P5UZW4 A0A0P5QT45 A0A0P5ANJ4 A0A0P5XLL3 A0A0N8D858 A0A0P5BPA1 A0A0P5S921 A0A0P5EVV4 A0A0P4WNJ2 A0A0N8B6J0 A0A0P4XSM5 A0A0P5NQE8 A0A0P5PQ66 A0A0P5TTV7 A0A0P6E2W6 A0A0N8CSI5 A0A0P5C0E8 A0A0P5J9C0 A0A0P6INE5 B7QJ27 A0A0P5RFN3 A0A0P5VXG0 A0A084WLA1 A0A0P5WMS4 A0A0P5NAK5 A0A0P5J3L4 A0A0P5J272 A0A0P6EEU7 A0A0P5SFR1 A0A0P5G0C7 A0A0P6IJI1 A0A0P6F6H1 A0A0P5C0B7
PDB
3LAF
E-value=3.27566e-21,
Score=250
Ontologies
GO
PANTHER
Topology
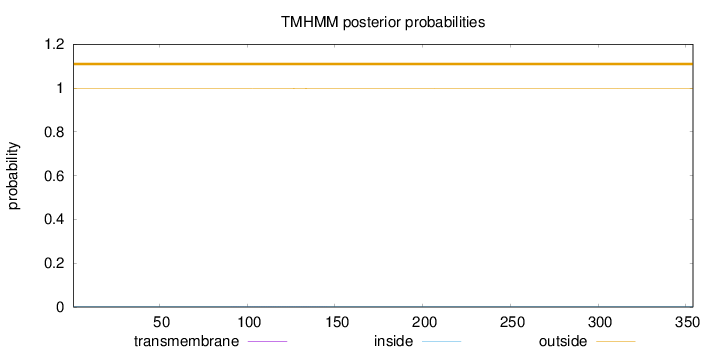
Length:
354
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01826
Exp number, first 60 AAs:
0.0008
Total prob of N-in:
0.00213
outside
1 - 354
Population Genetic Test Statistics
Pi
180.291001
Theta
195.600933
Tajima's D
-0.918352
CLR
1203.786304
CSRT
0.152892355382231
Interpretation
Uncertain
