Gene
KWMTBOMO00661
Pre Gene Modal
BGIBMGA003882
Annotation
PREDICTED:_proton-coupled_amino_acid_transporter_1-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.94
Sequence
CDS
ATGGACGAGAACAAAGCGGAGACGATGTACCTTCGTGCTGTCCCTGACGACGTCAACGGTGAAGCAAAGGACTCTGCAGCCGAAGAGAACTACGACCCTCACGAACATAGGCAACTGCCGAAGCCGACCAATAACATTGAGACCCTGATCCACCTGCTGAAATGTAGCCTGGGCACCGGCATCCTGGCCATGCCCCAGGCGTTCGCTCGCGCTGGCCTCGTCACCGGCATAGTGTTCACGGTGCTCATCGGAGTCCTGGTCACGCACTGCCTCCACGTGCTCGTGCGGTCGCAGTACGCGGCTTGCAAGCACCTCCGCGTGCCCCTGCTGTCGTACCCGGCCTCCATGGCGGCCGCGCTGGAGGTCGGCCCGGCTCCCTTCAGGAGGCTAGCCCGTCCCGCCTCTATCACCGTAGATATCTTCCTCGTTGTCTACCAGCTCGGCATCTGTTGTGTATACATCGTATTCATCGCGGACAACATCAAGAAGATCGTGGATCCGTTCTACGCGATGGCGGTGGAACTGCACATGCTGATCATCCTCTGCCCGTTAATCGTATTTAACCTGATCCCTAGTCTAAAACTGCTGGCGCCATTCTCAGCCGTGGCGAACGTACTCACATTCATAGGGCTCGGCATCGTGGTCTACTACCTGGCGACCGGCAAGAAGTCCCACCAGCCGCTCGACTTGTGGGGCTCGCTAGAAACCTTCCCGCTGTTCTTCGGCACCGTCTTGTTCGCGCTGACGGCGGTCGGTGTGGTTATAGCTCTGGAGAACAATATGAAGACTCCGAAGGCGTTCGGCAGCCCGTGCGGCGTGCTCAACTCGGGCATGGCGCTGATCGTGCTCTTGTACGTGACGGTGGGCGCCATGGGCTACATCTACTGCGTCTCCAAGTGCTCCGACTCCATCACTCTCGATCTTCCTTCCGGGCCGTTGGCGACTAGCGTCATCGTGATGTTCGCGGTGGCCATCTTCATTAGCTACGGACTGCACTGCTTCGTGCCGGTGGACGTGGTGTGGCGCGGGTACGTGGAGCCCCGCCTCCGAGCCGCGGCCACTCCGCCCGCGCGCCTCGTGCTCGCTGAATACGTGCTGCGCGTCCTGCTCTGTCTCCTCACATTCGTGCTGGCCGTGAGCGTGCCCCGCCTGGGACTGTTCATCTCTCTGTTCGGCGCGCTGTGCCTGTCGGCGCTGGGCATCTGTTTCCCGGCTATAATGGAAGCATGCGTGAAGTTCCCGCACGATCTGAAACCGATCTGGCTGTTGAAGGACGTACTGCTGTTTCTGGTGGGCATTGTGGGTCTGGTGGCCGGCACGTACACGGCTCTGCAGGCCATCTTCCGCTCGTTCCAGTCGACGAGCCCGCTCCACGCCTAA
Protein
MDENKAETMYLRAVPDDVNGEAKDSAAEENYDPHEHRQLPKPTNNIETLIHLLKCSLGTGILAMPQAFARAGLVTGIVFTVLIGVLVTHCLHVLVRSQYAACKHLRVPLLSYPASMAAALEVGPAPFRRLARPASITVDIFLVVYQLGICCVYIVFIADNIKKIVDPFYAMAVELHMLIILCPLIVFNLIPSLKLLAPFSAVANVLTFIGLGIVVYYLATGKKSHQPLDLWGSLETFPLFFGTVLFALTAVGVVIALENNMKTPKAFGSPCGVLNSGMALIVLLYVTVGAMGYIYCVSKCSDSITLDLPSGPLATSVIVMFAVAIFISYGLHCFVPVDVVWRGYVEPRLRAAATPPARLVLAEYVLRVLLCLLTFVLAVSVPRLGLFISLFGALCLSALGICFPAIMEACVKFPHDLKPIWLLKDVLLFLVGIVGLVAGTYTALQAIFRSFQSTSPLHA
Summary
Uniprot
H9J2Z5
A0A2W1BQK4
A0A2A4J0F9
A0A2H1W9E7
A0A212EKF0
A0A194QLH3
+ More
A0A194PFT1 A0A1Q3FER5 A0A1Q3FC12 B0WVQ8 A0A1Q3FCR9 A0A1B0D729 A0A1S4FFR9 A0A182KTL6 A0A182HRJ0 A0A182V2R5 A0A023ESI9 A0A182QGB4 A0A182W915 A0A182NAR5 A0A182KC28 A0A182IZS9 T1DU40 Q173C3 A0A182U6K1 A0A182RR72 A0A084W8K8 A0A182F3U9 W5JR06 A0A023FA30 R4FJW7 A0A1S4HF31 T1IBB4 A0A182YR04 A0A1L8DE49 A0A1L8DEB0 V9IJS5 A0A088AIY7 A0A0A9W4E0 A0A067RC98 A0A0M8ZPR3 A0A0L7R8G5 U5EUX7 E1ZXG5 E0VZX3 A0A2J7PW26 E2BM87 F4WKB5 A0A3L8DIK5 A0A0J7L2Z8 A0A158NQJ3 A0A2S2Q6J6 A0A154PQC2 A0A224XQH1 A0A182PCA8 A0A195EQQ6 A0A151XAU4 A0A1B6KMG1 A0A1Y1NIZ6 D6WF63 A0A1B6FW34 A0A1B6EVZ8 A0A1B6DVE7 A0A1Y1NDP8 K7JNJ2 A0A1J1I5J3 A0A2R7X7G0 A0A2H8TP20 A0A195BSP2 J9K7T3 A0A195D3R9 A0A151XAU2 N6T043 A0A2J7PW22 A0A2A3E666 A0A2A3E5F9 A0A088AIY5 E0VZX4 A0A067RLL7 A0A232FA51 A0A195DI13 A0A1S3DLC9 A0A3Q0JGM2 A0A0L7R8S2 A0A158NQR8 A0A026VX47 B4N441 A0A1L8DEQ7 A0A1Y0AWM7 A0A0M4EAI7 A0A1J1IZ53 A0A182RR73 A0A1Y1NHF4 A0A182W914 A0A182PCA9 B4KWF6 A0A182IUZ7 A0A0J7NX94 A0A084W8L0 Q7Q198
A0A194PFT1 A0A1Q3FER5 A0A1Q3FC12 B0WVQ8 A0A1Q3FCR9 A0A1B0D729 A0A1S4FFR9 A0A182KTL6 A0A182HRJ0 A0A182V2R5 A0A023ESI9 A0A182QGB4 A0A182W915 A0A182NAR5 A0A182KC28 A0A182IZS9 T1DU40 Q173C3 A0A182U6K1 A0A182RR72 A0A084W8K8 A0A182F3U9 W5JR06 A0A023FA30 R4FJW7 A0A1S4HF31 T1IBB4 A0A182YR04 A0A1L8DE49 A0A1L8DEB0 V9IJS5 A0A088AIY7 A0A0A9W4E0 A0A067RC98 A0A0M8ZPR3 A0A0L7R8G5 U5EUX7 E1ZXG5 E0VZX3 A0A2J7PW26 E2BM87 F4WKB5 A0A3L8DIK5 A0A0J7L2Z8 A0A158NQJ3 A0A2S2Q6J6 A0A154PQC2 A0A224XQH1 A0A182PCA8 A0A195EQQ6 A0A151XAU4 A0A1B6KMG1 A0A1Y1NIZ6 D6WF63 A0A1B6FW34 A0A1B6EVZ8 A0A1B6DVE7 A0A1Y1NDP8 K7JNJ2 A0A1J1I5J3 A0A2R7X7G0 A0A2H8TP20 A0A195BSP2 J9K7T3 A0A195D3R9 A0A151XAU2 N6T043 A0A2J7PW22 A0A2A3E666 A0A2A3E5F9 A0A088AIY5 E0VZX4 A0A067RLL7 A0A232FA51 A0A195DI13 A0A1S3DLC9 A0A3Q0JGM2 A0A0L7R8S2 A0A158NQR8 A0A026VX47 B4N441 A0A1L8DEQ7 A0A1Y0AWM7 A0A0M4EAI7 A0A1J1IZ53 A0A182RR73 A0A1Y1NHF4 A0A182W914 A0A182PCA9 B4KWF6 A0A182IUZ7 A0A0J7NX94 A0A084W8L0 Q7Q198
Pubmed
EMBL
BABH01031182
KZ149976
PZC75914.1
NWSH01003944
PCG65647.1
ODYU01007156
+ More
SOQ49690.1 AGBW02014274 OWR41963.1 KQ461196 KPJ06408.1 KQ459605 KPI91888.1 GFDL01009022 JAV26023.1 GFDL01009924 JAV25121.1 DS232131 EDS35665.1 GFDL01009654 JAV25391.1 AJVK01012301 AJVK01012302 AJVK01012303 APCN01001708 APCN01001709 GAPW01001377 GAPW01001376 JAC12222.1 AXCN02000660 GAMD01000484 JAB01107.1 CH477427 CH477217 EAT41137.1 EAT47448.1 ATLV01021488 KE525319 KFB46552.1 ADMH02000680 ETN65335.1 GBBI01000858 JAC17854.1 GAHY01002113 JAA75397.1 AAAB01008980 ACPB03000217 GFDF01009366 JAV04718.1 GFDF01009367 JAV04717.1 JR049241 AEY60907.1 GBHO01040955 GBRD01013400 GBRD01011200 GBRD01011199 GBRD01011198 GBRD01011196 GBRD01011195 GBRD01011194 GBRD01011193 GBRD01010661 GBRD01010660 GBRD01010659 GDHC01013528 GDHC01010376 GDHC01006795 JAG02649.1 JAG52426.1 JAQ05101.1 JAQ08253.1 JAQ11834.1 KK852550 KDR21506.1 KQ435922 KOX68585.1 KQ414632 KOC67165.1 GANO01002132 JAB57739.1 GL435038 EFN74142.1 DS235854 EEB18928.1 NEVH01020938 PNF20534.1 GL449158 EFN83213.1 GL888199 EGI65339.1 QOIP01000008 RLU19973.1 LBMM01001038 KMQ97036.1 ADTU01023344 GGMS01004018 MBY73221.1 KQ434998 KZC13310.1 GFTR01006033 JAW10393.1 KQ982021 KYN30543.1 KQ982335 KYQ57503.1 GEBQ01027345 JAT12632.1 GEZM01005587 GEZM01005583 JAV96076.1 KQ971327 EEZ99836.2 GECZ01015369 JAS54400.1 GECZ01027691 JAS42078.1 GEDC01026267 GEDC01017056 GEDC01008676 GEDC01008271 GEDC01007642 GEDC01005657 GEDC01003083 JAS11031.1 JAS20242.1 JAS28622.1 JAS29027.1 JAS29656.1 JAS31641.1 JAS34215.1 GEZM01005585 GEZM01005581 JAV96074.1 CVRI01000042 CRK95552.1 KK857638 PTY27370.1 GFXV01003964 MBW15769.1 KQ976419 KYM89315.1 ABLF02029405 ABLF02029406 ABLF02029409 KQ976885 KYN07555.1 KYQ57502.1 APGK01057331 APGK01057332 KB741280 KB631604 ENN70873.1 ERL84593.1 PNF20533.1 KZ288372 PBC26726.1 PBC26724.1 EEB18929.1 KDR21505.1 NNAY01000622 OXU27339.1 KQ980824 KYN12477.1 KOC67166.1 ADTU01023342 ADTU01023343 KK107648 EZA48332.1 RLU19972.1 CH964095 EDW78915.1 GFDF01009158 JAV04926.1 KY921804 ART29393.1 CP012525 ALC44278.1 CVRI01000063 CRL04446.1 GEZM01005586 GEZM01005584 JAV96075.1 CH933809 EDW19585.1 KRG06698.1 KMQ97035.1 ATLV01021489 KFB46554.1 EAA14378.3
SOQ49690.1 AGBW02014274 OWR41963.1 KQ461196 KPJ06408.1 KQ459605 KPI91888.1 GFDL01009022 JAV26023.1 GFDL01009924 JAV25121.1 DS232131 EDS35665.1 GFDL01009654 JAV25391.1 AJVK01012301 AJVK01012302 AJVK01012303 APCN01001708 APCN01001709 GAPW01001377 GAPW01001376 JAC12222.1 AXCN02000660 GAMD01000484 JAB01107.1 CH477427 CH477217 EAT41137.1 EAT47448.1 ATLV01021488 KE525319 KFB46552.1 ADMH02000680 ETN65335.1 GBBI01000858 JAC17854.1 GAHY01002113 JAA75397.1 AAAB01008980 ACPB03000217 GFDF01009366 JAV04718.1 GFDF01009367 JAV04717.1 JR049241 AEY60907.1 GBHO01040955 GBRD01013400 GBRD01011200 GBRD01011199 GBRD01011198 GBRD01011196 GBRD01011195 GBRD01011194 GBRD01011193 GBRD01010661 GBRD01010660 GBRD01010659 GDHC01013528 GDHC01010376 GDHC01006795 JAG02649.1 JAG52426.1 JAQ05101.1 JAQ08253.1 JAQ11834.1 KK852550 KDR21506.1 KQ435922 KOX68585.1 KQ414632 KOC67165.1 GANO01002132 JAB57739.1 GL435038 EFN74142.1 DS235854 EEB18928.1 NEVH01020938 PNF20534.1 GL449158 EFN83213.1 GL888199 EGI65339.1 QOIP01000008 RLU19973.1 LBMM01001038 KMQ97036.1 ADTU01023344 GGMS01004018 MBY73221.1 KQ434998 KZC13310.1 GFTR01006033 JAW10393.1 KQ982021 KYN30543.1 KQ982335 KYQ57503.1 GEBQ01027345 JAT12632.1 GEZM01005587 GEZM01005583 JAV96076.1 KQ971327 EEZ99836.2 GECZ01015369 JAS54400.1 GECZ01027691 JAS42078.1 GEDC01026267 GEDC01017056 GEDC01008676 GEDC01008271 GEDC01007642 GEDC01005657 GEDC01003083 JAS11031.1 JAS20242.1 JAS28622.1 JAS29027.1 JAS29656.1 JAS31641.1 JAS34215.1 GEZM01005585 GEZM01005581 JAV96074.1 CVRI01000042 CRK95552.1 KK857638 PTY27370.1 GFXV01003964 MBW15769.1 KQ976419 KYM89315.1 ABLF02029405 ABLF02029406 ABLF02029409 KQ976885 KYN07555.1 KYQ57502.1 APGK01057331 APGK01057332 KB741280 KB631604 ENN70873.1 ERL84593.1 PNF20533.1 KZ288372 PBC26726.1 PBC26724.1 EEB18929.1 KDR21505.1 NNAY01000622 OXU27339.1 KQ980824 KYN12477.1 KOC67166.1 ADTU01023342 ADTU01023343 KK107648 EZA48332.1 RLU19972.1 CH964095 EDW78915.1 GFDF01009158 JAV04926.1 KY921804 ART29393.1 CP012525 ALC44278.1 CVRI01000063 CRL04446.1 GEZM01005586 GEZM01005584 JAV96075.1 CH933809 EDW19585.1 KRG06698.1 KMQ97035.1 ATLV01021489 KFB46554.1 EAA14378.3
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000002320
+ More
UP000092462 UP000075882 UP000075840 UP000075903 UP000075886 UP000075920 UP000075884 UP000075881 UP000075880 UP000008820 UP000075902 UP000075900 UP000030765 UP000069272 UP000000673 UP000015103 UP000076408 UP000005203 UP000027135 UP000053105 UP000053825 UP000000311 UP000009046 UP000235965 UP000008237 UP000007755 UP000279307 UP000036403 UP000005205 UP000076502 UP000075885 UP000078541 UP000075809 UP000007266 UP000002358 UP000183832 UP000078540 UP000007819 UP000078542 UP000019118 UP000030742 UP000242457 UP000215335 UP000078492 UP000079169 UP000053097 UP000007798 UP000092553 UP000009192 UP000007062
UP000092462 UP000075882 UP000075840 UP000075903 UP000075886 UP000075920 UP000075884 UP000075881 UP000075880 UP000008820 UP000075902 UP000075900 UP000030765 UP000069272 UP000000673 UP000015103 UP000076408 UP000005203 UP000027135 UP000053105 UP000053825 UP000000311 UP000009046 UP000235965 UP000008237 UP000007755 UP000279307 UP000036403 UP000005205 UP000076502 UP000075885 UP000078541 UP000075809 UP000007266 UP000002358 UP000183832 UP000078540 UP000007819 UP000078542 UP000019118 UP000030742 UP000242457 UP000215335 UP000078492 UP000079169 UP000053097 UP000007798 UP000092553 UP000009192 UP000007062
PRIDE
Pfam
PF01490 Aa_trans
SUPFAM
SSF57903
SSF57903
ProteinModelPortal
H9J2Z5
A0A2W1BQK4
A0A2A4J0F9
A0A2H1W9E7
A0A212EKF0
A0A194QLH3
+ More
A0A194PFT1 A0A1Q3FER5 A0A1Q3FC12 B0WVQ8 A0A1Q3FCR9 A0A1B0D729 A0A1S4FFR9 A0A182KTL6 A0A182HRJ0 A0A182V2R5 A0A023ESI9 A0A182QGB4 A0A182W915 A0A182NAR5 A0A182KC28 A0A182IZS9 T1DU40 Q173C3 A0A182U6K1 A0A182RR72 A0A084W8K8 A0A182F3U9 W5JR06 A0A023FA30 R4FJW7 A0A1S4HF31 T1IBB4 A0A182YR04 A0A1L8DE49 A0A1L8DEB0 V9IJS5 A0A088AIY7 A0A0A9W4E0 A0A067RC98 A0A0M8ZPR3 A0A0L7R8G5 U5EUX7 E1ZXG5 E0VZX3 A0A2J7PW26 E2BM87 F4WKB5 A0A3L8DIK5 A0A0J7L2Z8 A0A158NQJ3 A0A2S2Q6J6 A0A154PQC2 A0A224XQH1 A0A182PCA8 A0A195EQQ6 A0A151XAU4 A0A1B6KMG1 A0A1Y1NIZ6 D6WF63 A0A1B6FW34 A0A1B6EVZ8 A0A1B6DVE7 A0A1Y1NDP8 K7JNJ2 A0A1J1I5J3 A0A2R7X7G0 A0A2H8TP20 A0A195BSP2 J9K7T3 A0A195D3R9 A0A151XAU2 N6T043 A0A2J7PW22 A0A2A3E666 A0A2A3E5F9 A0A088AIY5 E0VZX4 A0A067RLL7 A0A232FA51 A0A195DI13 A0A1S3DLC9 A0A3Q0JGM2 A0A0L7R8S2 A0A158NQR8 A0A026VX47 B4N441 A0A1L8DEQ7 A0A1Y0AWM7 A0A0M4EAI7 A0A1J1IZ53 A0A182RR73 A0A1Y1NHF4 A0A182W914 A0A182PCA9 B4KWF6 A0A182IUZ7 A0A0J7NX94 A0A084W8L0 Q7Q198
A0A194PFT1 A0A1Q3FER5 A0A1Q3FC12 B0WVQ8 A0A1Q3FCR9 A0A1B0D729 A0A1S4FFR9 A0A182KTL6 A0A182HRJ0 A0A182V2R5 A0A023ESI9 A0A182QGB4 A0A182W915 A0A182NAR5 A0A182KC28 A0A182IZS9 T1DU40 Q173C3 A0A182U6K1 A0A182RR72 A0A084W8K8 A0A182F3U9 W5JR06 A0A023FA30 R4FJW7 A0A1S4HF31 T1IBB4 A0A182YR04 A0A1L8DE49 A0A1L8DEB0 V9IJS5 A0A088AIY7 A0A0A9W4E0 A0A067RC98 A0A0M8ZPR3 A0A0L7R8G5 U5EUX7 E1ZXG5 E0VZX3 A0A2J7PW26 E2BM87 F4WKB5 A0A3L8DIK5 A0A0J7L2Z8 A0A158NQJ3 A0A2S2Q6J6 A0A154PQC2 A0A224XQH1 A0A182PCA8 A0A195EQQ6 A0A151XAU4 A0A1B6KMG1 A0A1Y1NIZ6 D6WF63 A0A1B6FW34 A0A1B6EVZ8 A0A1B6DVE7 A0A1Y1NDP8 K7JNJ2 A0A1J1I5J3 A0A2R7X7G0 A0A2H8TP20 A0A195BSP2 J9K7T3 A0A195D3R9 A0A151XAU2 N6T043 A0A2J7PW22 A0A2A3E666 A0A2A3E5F9 A0A088AIY5 E0VZX4 A0A067RLL7 A0A232FA51 A0A195DI13 A0A1S3DLC9 A0A3Q0JGM2 A0A0L7R8S2 A0A158NQR8 A0A026VX47 B4N441 A0A1L8DEQ7 A0A1Y0AWM7 A0A0M4EAI7 A0A1J1IZ53 A0A182RR73 A0A1Y1NHF4 A0A182W914 A0A182PCA9 B4KWF6 A0A182IUZ7 A0A0J7NX94 A0A084W8L0 Q7Q198
PDB
6C08
E-value=0.0305957,
Score=88
Ontologies
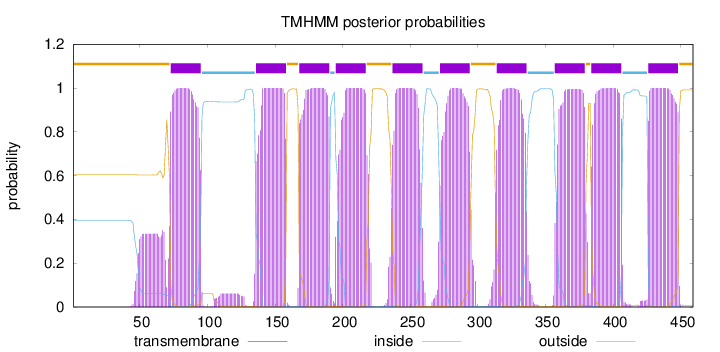
Topology
Length:
459
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
232.50086
Exp number, first 60 AAs:
4.24999
Total prob of N-in:
0.39640
outside
1 - 72
TMhelix
73 - 95
inside
96 - 135
TMhelix
136 - 158
outside
159 - 167
TMhelix
168 - 190
inside
191 - 194
TMhelix
195 - 217
outside
218 - 236
TMhelix
237 - 259
inside
260 - 271
TMhelix
272 - 294
outside
295 - 313
TMhelix
314 - 336
inside
337 - 356
TMhelix
357 - 379
outside
380 - 383
TMhelix
384 - 406
inside
407 - 425
TMhelix
426 - 448
outside
449 - 459
Population Genetic Test Statistics
Pi
3.304942
Theta
16.893238
Tajima's D
-1.400563
CLR
0.177665
CSRT
0.072396380180991
Interpretation
Uncertain
