Gene
KWMTBOMO00659
Pre Gene Modal
BGIBMGA003883
Annotation
PREDICTED:_piggyBac_transposable_element-derived_protein_3-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.572 Nuclear Reliability : 2.095
Sequence
CDS
ATGCCGCGATTACTACAAAGCGAAATTATAAAGTATTTAGAAACTCTTGAAGATTCAGAAGATGGCTTAGACGGGTCAGACTCCGAAGTAGAGGATGAAGGTCACTATTATCAAACAGCTCGAGATGTTGTTCGAGACCTTGAAAATAGTGATGAAGGTGGAATGCCCAGTCTGATGAAGTGGATGCTGATCCTCCTCTCATTTGAGGTTTACAGTGGAGGGGGCTCCAAAAATAACATCTTACCTGGTGAGCCAGATTTGGGAGAGTCTGGCAATACAGTCGTCAGGTTAGCTAGAATGGTACCACGCAATGTGAATCACATTATATACTTCGATAACTTCTATACTTCTGTGCCTCTCGTCACCTATTTAGCTAAGGAAGGAATTTTCTCACTGGGAACAGTAAGAGTAAACCGATTGAGAAACTGCAAATTGCCTGAAAAAAGCACTATAATTAAAAAGAACGTACCCAGAGGATTCTATGAGGAAAACGTGGCGACTGTGGATGATATTGACGTAAGTGCTGTTGTGTGGAAAGATAACAAGCCCGTTAACCTTCTTTCCACTTACGTAGGAGCAAAACCAGCAATCACAGTCACACGCTTTGATAAATCGAAGAAAGAGAGAGTTGCTATTCCATGCCCCAAAGTCATCAAGGATTACAATTCCCACATGGGAGGCGTCGACTTATTAGATTCATTTATAGGTCGTTATCGCATCACAATGAAGAGCCGCAAGTGGACCATGCGCTTATTTTATCATTTCCTGGACCTCGCAGTTATAAATTCATGGGTTATGTTCAAGAAGGTAAATAATATAAAAGGAAATGATCAACTTCTCAATTTGGGTACATTCAGACTAGAACTTGCTGAAACCCTCTGCAAATTGGGTCTACCTGCAAATGGCGCTAAACGTGGTCGACCTAGTGGGAGCACAATACAAAGAGAACTTGTAATGAAGAAGTTCCGAGGACCAGCTCAAGCAATTCCTTTGAAAGACGTCAGATTGGATCAGACATGTCATTGGGCAGTTTGGCTGGATAAGCAGCAGAGGTGCAAATTCCCAAAATGTTCAGGCTTTACATTCAAAATGTGCCAAAAATGTCGTGTATCTTTATGTGATACGAAAAAAAGCAATTGTTTTTATAAATATCATAATGAAGAATAA
Protein
MPRLLQSEIIKYLETLEDSEDGLDGSDSEVEDEGHYYQTARDVVRDLENSDEGGMPSLMKWMLILLSFEVYSGGGSKNNILPGEPDLGESGNTVVRLARMVPRNVNHIIYFDNFYTSVPLVTYLAKEGIFSLGTVRVNRLRNCKLPEKSTIIKKNVPRGFYEENVATVDDIDVSAVVWKDNKPVNLLSTYVGAKPAITVTRFDKSKKERVAIPCPKVIKDYNSHMGGVDLLDSFIGRYRITMKSRKWTMRLFYHFLDLAVINSWVMFKKVNNIKGNDQLLNLGTFRLELAETLCKLGLPANGAKRGRPSGSTIQRELVMKKFRGPAQAIPLKDVRLDQTCHWAVWLDKQQRCKFPKCSGFTFKMCQKCRVSLCDTKKSNCFYKYHNEE
Summary
Uniprot
H9J2Z6
A0A2H1VR69
A0A2H1WTL9
H9JGZ6
H9JXX7
S4UKH5
+ More
A0A087T4S1 A0A1E1WQY2 A0A2S2PBK5 A0A212EHM2 X1X2F2 J9LW60 A0A2H8TUQ3 A0A212FFX4 A0A3B3BTP2 A0A0A1WEM0 A0A146TTD4 A0A3P9K060 A0A3P9ITA5 A0A146R6K8 A0A1B6I2X1 A0A1B6K7Q3 A0A2H8TNX6 A0A3B3I560 J9M9C5 V5HIC5 A0A3P8SS87 A0A3B5R7A5 A0A3P9J3H4 A0A3B3I2Y1 A0A224YVZ2 A0A3B3IL37 A0A3B3I0J3 A0A3P9HE00 A0A3B3H5M7 J9KQV2 T1HDP7 A0A3P9I3M3 A0A3B3SQ04 A0A3P9LFA7 A0A3B3HE36 A0A3B3IE75 A0A3B5QXE4 A0A3P9IAA1 H2L551 A0A3B4ZMF2 A0A3P9LG76 A0A1B6MG92 A0A0P4W1S1 A0A146SI21 A0A3P9LX47 A0A3B3RJN9 A0A147BC68 H2T2T7 A0A3B3S9X6 A0A146PY66 V5HDK4 A0A3P9JUS2 A0A0K0EC24 A0A147BJS4 A0A3B3TU82 A0A3P9NMX7 A0A3B3XUL4 A0A090X7K4 A0A3B3TB08 A0A315UTZ6 A0A0S7MA33 M3ZMJ7 A0A3P9PHM0 A0A3P9PHH9 A0A3B3UK40 A0A3B3YVN1 A0A146S7X5 A0A3B4EUY8 A0A3B1KH96 A0A3P9IE40 A0A096M1J4 A0A3B4A161 A0A146VPP3 A0A147BBQ2 A0A3P9I8W3 T2MIT5 A0A087Y5J4 A0A3B5N0U8 A0A2S2NW57 A0A2H1X084 A0A147BJU4 A0A2J7Q3F6
A0A087T4S1 A0A1E1WQY2 A0A2S2PBK5 A0A212EHM2 X1X2F2 J9LW60 A0A2H8TUQ3 A0A212FFX4 A0A3B3BTP2 A0A0A1WEM0 A0A146TTD4 A0A3P9K060 A0A3P9ITA5 A0A146R6K8 A0A1B6I2X1 A0A1B6K7Q3 A0A2H8TNX6 A0A3B3I560 J9M9C5 V5HIC5 A0A3P8SS87 A0A3B5R7A5 A0A3P9J3H4 A0A3B3I2Y1 A0A224YVZ2 A0A3B3IL37 A0A3B3I0J3 A0A3P9HE00 A0A3B3H5M7 J9KQV2 T1HDP7 A0A3P9I3M3 A0A3B3SQ04 A0A3P9LFA7 A0A3B3HE36 A0A3B3IE75 A0A3B5QXE4 A0A3P9IAA1 H2L551 A0A3B4ZMF2 A0A3P9LG76 A0A1B6MG92 A0A0P4W1S1 A0A146SI21 A0A3P9LX47 A0A3B3RJN9 A0A147BC68 H2T2T7 A0A3B3S9X6 A0A146PY66 V5HDK4 A0A3P9JUS2 A0A0K0EC24 A0A147BJS4 A0A3B3TU82 A0A3P9NMX7 A0A3B3XUL4 A0A090X7K4 A0A3B3TB08 A0A315UTZ6 A0A0S7MA33 M3ZMJ7 A0A3P9PHM0 A0A3P9PHH9 A0A3B3UK40 A0A3B3YVN1 A0A146S7X5 A0A3B4EUY8 A0A3B1KH96 A0A3P9IE40 A0A096M1J4 A0A3B4A161 A0A146VPP3 A0A147BBQ2 A0A3P9I8W3 T2MIT5 A0A087Y5J4 A0A3B5N0U8 A0A2S2NW57 A0A2H1X084 A0A147BJU4 A0A2J7Q3F6
Pubmed
EMBL
BABH01031190
ODYU01003952
SOQ43341.1
ODYU01010930
SOQ56347.1
BABH01037818
+ More
BABH01040586 BABH01040587 BABH01040588 JX392388 AGK25051.1 KK113395 KFM60110.1 GDQN01001619 JAT89435.1 GGMR01014224 MBY26843.1 AGBW02014856 OWR40986.1 ABLF02058363 ABLF02066746 ABLF02010159 GFXV01005864 MBW17669.1 AGBW02008750 OWR52641.1 GBXI01016833 JAC97458.1 GCES01090325 JAQ95997.1 GCES01122712 JAQ63610.1 GECU01026434 JAS81272.1 GECU01000219 JAT07488.1 GFXV01003527 MBW15332.1 ABLF02009142 GANP01011210 JAB73258.1 GFPF01010630 MAA21776.1 ABLF02009898 ACPB03028099 GEBQ01005051 JAT34926.1 GDRN01091243 GDRN01091242 JAI60307.1 GCES01106053 JAQ80269.1 GEGO01007352 JAR88052.1 GCES01137554 JAQ48768.1 GANP01011226 JAB73242.1 GEGO01004386 JAR91018.1 GBIH01002643 JAC92067.1 NHOQ01002733 PWA15177.1 GBYX01091460 JAO98581.1 GCES01109695 JAQ76627.1 AYCK01018182 GCES01066870 JAR19453.1 GEGO01007223 JAR88181.1 HAAD01005618 CDG71850.1 AYCK01002766 GGMR01008842 MBY21461.1 ODYU01012425 SOQ58723.1 GEGO01004410 JAR90994.1 NEVH01019067 PNF23115.1
BABH01040586 BABH01040587 BABH01040588 JX392388 AGK25051.1 KK113395 KFM60110.1 GDQN01001619 JAT89435.1 GGMR01014224 MBY26843.1 AGBW02014856 OWR40986.1 ABLF02058363 ABLF02066746 ABLF02010159 GFXV01005864 MBW17669.1 AGBW02008750 OWR52641.1 GBXI01016833 JAC97458.1 GCES01090325 JAQ95997.1 GCES01122712 JAQ63610.1 GECU01026434 JAS81272.1 GECU01000219 JAT07488.1 GFXV01003527 MBW15332.1 ABLF02009142 GANP01011210 JAB73258.1 GFPF01010630 MAA21776.1 ABLF02009898 ACPB03028099 GEBQ01005051 JAT34926.1 GDRN01091243 GDRN01091242 JAI60307.1 GCES01106053 JAQ80269.1 GEGO01007352 JAR88052.1 GCES01137554 JAQ48768.1 GANP01011226 JAB73242.1 GEGO01004386 JAR91018.1 GBIH01002643 JAC92067.1 NHOQ01002733 PWA15177.1 GBYX01091460 JAO98581.1 GCES01109695 JAQ76627.1 AYCK01018182 GCES01066870 JAR19453.1 GEGO01007223 JAR88181.1 HAAD01005618 CDG71850.1 AYCK01002766 GGMR01008842 MBY21461.1 ODYU01012425 SOQ58723.1 GEGO01004410 JAR90994.1 NEVH01019067 PNF23115.1
Proteomes
UP000005204
UP000054359
UP000007151
UP000007819
UP000261560
UP000265180
+ More
UP000265200 UP000001038 UP000265080 UP000002852 UP000015103 UP000261540 UP000261400 UP000265000 UP000005226 UP000035681 UP000261500 UP000242638 UP000261480 UP000261460 UP000018467 UP000028760 UP000261520 UP000261380 UP000235965
UP000265200 UP000001038 UP000265080 UP000002852 UP000015103 UP000261540 UP000261400 UP000265000 UP000005226 UP000035681 UP000261500 UP000242638 UP000261480 UP000261460 UP000018467 UP000028760 UP000261520 UP000261380 UP000235965
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9J2Z6
A0A2H1VR69
A0A2H1WTL9
H9JGZ6
H9JXX7
S4UKH5
+ More
A0A087T4S1 A0A1E1WQY2 A0A2S2PBK5 A0A212EHM2 X1X2F2 J9LW60 A0A2H8TUQ3 A0A212FFX4 A0A3B3BTP2 A0A0A1WEM0 A0A146TTD4 A0A3P9K060 A0A3P9ITA5 A0A146R6K8 A0A1B6I2X1 A0A1B6K7Q3 A0A2H8TNX6 A0A3B3I560 J9M9C5 V5HIC5 A0A3P8SS87 A0A3B5R7A5 A0A3P9J3H4 A0A3B3I2Y1 A0A224YVZ2 A0A3B3IL37 A0A3B3I0J3 A0A3P9HE00 A0A3B3H5M7 J9KQV2 T1HDP7 A0A3P9I3M3 A0A3B3SQ04 A0A3P9LFA7 A0A3B3HE36 A0A3B3IE75 A0A3B5QXE4 A0A3P9IAA1 H2L551 A0A3B4ZMF2 A0A3P9LG76 A0A1B6MG92 A0A0P4W1S1 A0A146SI21 A0A3P9LX47 A0A3B3RJN9 A0A147BC68 H2T2T7 A0A3B3S9X6 A0A146PY66 V5HDK4 A0A3P9JUS2 A0A0K0EC24 A0A147BJS4 A0A3B3TU82 A0A3P9NMX7 A0A3B3XUL4 A0A090X7K4 A0A3B3TB08 A0A315UTZ6 A0A0S7MA33 M3ZMJ7 A0A3P9PHM0 A0A3P9PHH9 A0A3B3UK40 A0A3B3YVN1 A0A146S7X5 A0A3B4EUY8 A0A3B1KH96 A0A3P9IE40 A0A096M1J4 A0A3B4A161 A0A146VPP3 A0A147BBQ2 A0A3P9I8W3 T2MIT5 A0A087Y5J4 A0A3B5N0U8 A0A2S2NW57 A0A2H1X084 A0A147BJU4 A0A2J7Q3F6
A0A087T4S1 A0A1E1WQY2 A0A2S2PBK5 A0A212EHM2 X1X2F2 J9LW60 A0A2H8TUQ3 A0A212FFX4 A0A3B3BTP2 A0A0A1WEM0 A0A146TTD4 A0A3P9K060 A0A3P9ITA5 A0A146R6K8 A0A1B6I2X1 A0A1B6K7Q3 A0A2H8TNX6 A0A3B3I560 J9M9C5 V5HIC5 A0A3P8SS87 A0A3B5R7A5 A0A3P9J3H4 A0A3B3I2Y1 A0A224YVZ2 A0A3B3IL37 A0A3B3I0J3 A0A3P9HE00 A0A3B3H5M7 J9KQV2 T1HDP7 A0A3P9I3M3 A0A3B3SQ04 A0A3P9LFA7 A0A3B3HE36 A0A3B3IE75 A0A3B5QXE4 A0A3P9IAA1 H2L551 A0A3B4ZMF2 A0A3P9LG76 A0A1B6MG92 A0A0P4W1S1 A0A146SI21 A0A3P9LX47 A0A3B3RJN9 A0A147BC68 H2T2T7 A0A3B3S9X6 A0A146PY66 V5HDK4 A0A3P9JUS2 A0A0K0EC24 A0A147BJS4 A0A3B3TU82 A0A3P9NMX7 A0A3B3XUL4 A0A090X7K4 A0A3B3TB08 A0A315UTZ6 A0A0S7MA33 M3ZMJ7 A0A3P9PHM0 A0A3P9PHH9 A0A3B3UK40 A0A3B3YVN1 A0A146S7X5 A0A3B4EUY8 A0A3B1KH96 A0A3P9IE40 A0A096M1J4 A0A3B4A161 A0A146VPP3 A0A147BBQ2 A0A3P9I8W3 T2MIT5 A0A087Y5J4 A0A3B5N0U8 A0A2S2NW57 A0A2H1X084 A0A147BJU4 A0A2J7Q3F6
Ontologies
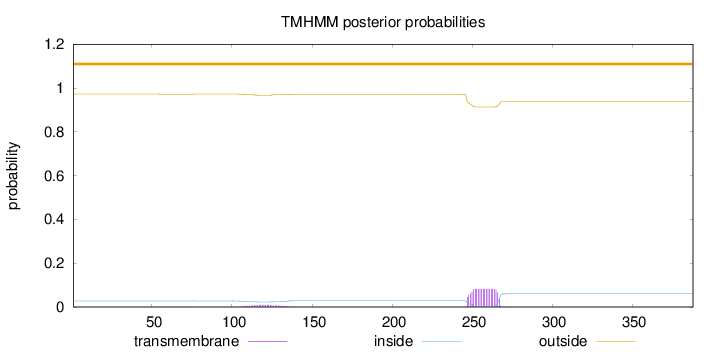
Topology
Length:
388
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.80963
Exp number, first 60 AAs:
0.00145
Total prob of N-in:
0.02769
outside
1 - 388
Population Genetic Test Statistics
Pi
16.71469
Theta
21.016982
Tajima's D
-0.593798
CLR
0.022306
CSRT
0.218539073046348
Interpretation
Uncertain
