Gene
KWMTBOMO00652
Pre Gene Modal
BGIBMGA003867
Annotation
PREDICTED:_G-protein-signaling_modulator_2_isoform_X1_[Amyelois_transitella]
Full name
S-adenosylmethionine sensor upstream of mTORC1
Alternative Name
Probable methyltransferase BMT2 homolog
Location in the cell
Cytoplasmic Reliability : 1.718 Nuclear Reliability : 2.098
Sequence
CDS
ATGTGTCTCGAGCTCGCTCTCGAAGGTGAGAGGCTCTGCAAGGCTGGCGACTGCAAAGCAGGAGTAGCCTTCTTCCAAGCGGCGATACAGGCGGGAACAGATGACCTACGCACCCTGAGCGCAATATACAGCCAAATGGGCAACGCTTATTTTTACCTTGGTGAATACCATAAAGCAGTGCAGTACCACCGACACGATCTAACTTTAGCAAGAACCATGAATGATAAGCTGGGCGAAGCCAAAGCTTCAGGTAATCTAGGCAACACCCTTAAAGTAATAGGAAAGTTCAATGAGGCCATACAGTGCTGCCTCCGACACCTGGAGATATCTCGCTCCCTGGGCGACAGAAATAGCGAGGGTAGAGCTCTGTACAACCTGGCCAACGTATACCATGCCCAAGGTAAACATCTGGGAAAGGTCGGACAAAACGATCCGGGACATTTTCCCCAAGAGGTGCGCGAATGCCTTTTACAAGCAGTCAACTATTATGAACAAAATTTGGTACTCATGCGTGAGCTGAACGATCGCCTAGCCATCGGTCGGGCTTGCGGCAACCTTGGAAACACTTACTACCTGCTTGGTGAATTTAATGTAGCTGTTCAGTACCACACGGAGCGACTGGAAATGGCCAAGGAATGTAAAGACAAGGCAGCCGAAAGACGCGCCCACAGCAATCTTGGAAACTCACACATTTTCCTCGGGCAGTTTGAGAACGCGGCCGATCATTACAAACAGACTTTGGCGATTGCCGAAGAATTAAAAGACGAAGCGGTGGTCGCACAAGCCTGCTACTCCTTAGGGAATACGTACACGTTACTCCGGGAGTATAGAATTGCGGAAGAGTACCATGCTAGGCACCTGGAGGCGGCCAGGAAGCTTCAGGATCGAGTTGGCGAAGGCAGGGCCTGTTGGTCTCTGGGCAACGCCCACGCGGCTCTCGGCAACCACGAGAAAGCATTGTATTATGCCATTGAACATTATACCATATCGAAAGAGATCTGTGATGTGCTGGGGATGGCCACGGCACAGATGAACATCAACGACCTCCGCAAGGTGCTCGGGCTCCCGCCGCTCGCCGAGACCGCCGCAGCTCAGCCCGCCCCGCCAGACCAAGCCGGCTCCAGCCAGACGCCCTTCAACGCCCTTCGTGTGAGGAGGCAGAGCATGGAACAGCTGTCCCTGATACAATTGACACCCGACGGTAAGAAGGACCCCGAGGACGCCCAGAAGAAAGCCGTGCTGATGCGTTCGGAATCCGAAGACAGCCCCATGGAGTCCTTCCTCGATCTGCTATCCAAATACCAATCCGAGAGGATGGATGAGCAGCGAGCCACCCTCGTGGAGACCAAAGAAATCCAAGAGACTCCCAAACCAACTGAGAAGCCGAAGCCCAGCTCCAGCAAACTGTTCCGCTCTTCCAGCAGCGGCAGCAAGCAGTACGATTCGAACGAGGCGCTCTTGGACATGATCACCGATCTCCAGTCCGAGCGCATGGACTCCCAGCGCGCCGAGCTGCGCCCGATCAAAAAGAAGCTGTCCAAGGAAATGAAGGGGACCTCGCTACCCGGCCTCCGCACCGCCCGCGTCGCCGGCGCCAAGCCGGAGGACGATTTCCTCGACATGATTGCTCGTGTACAGGGCACCCGGCTCGACGATCAACGGACCGAGATGCCTCGTCCCAAGGCCGACCCTCGTCCCAAGACCGTGCCCGATGACGATTTCTTCGCTCTGATCATGAGGGCCCAGTCTGGACGTATGGAGGACCAACGAGCTGCCATTCCGAAGCCGGCCGAGAACCGACAGAGCAAGCCGGCCCCCAAGCCGCGTAAATAG
Protein
MCLELALEGERLCKAGDCKAGVAFFQAAIQAGTDDLRTLSAIYSQMGNAYFYLGEYHKAVQYHRHDLTLARTMNDKLGEAKASGNLGNTLKVIGKFNEAIQCCLRHLEISRSLGDRNSEGRALYNLANVYHAQGKHLGKVGQNDPGHFPQEVRECLLQAVNYYEQNLVLMRELNDRLAIGRACGNLGNTYYLLGEFNVAVQYHTERLEMAKECKDKAAERRAHSNLGNSHIFLGQFENAADHYKQTLAIAEELKDEAVVAQACYSLGNTYTLLREYRIAEEYHARHLEAARKLQDRVGEGRACWSLGNAHAALGNHEKALYYAIEHYTISKEICDVLGMATAQMNINDLRKVLGLPPLAETAAAQPAPPDQAGSSQTPFNALRVRRQSMEQLSLIQLTPDGKKDPEDAQKKAVLMRSESEDSPMESFLDLLSKYQSERMDEQRATLVETKEIQETPKPTEKPKPSSSKLFRSSSSGSKQYDSNEALLDMITDLQSERMDSQRAELRPIKKKLSKEMKGTSLPGLRTARVAGAKPEDDFLDMIARVQGTRLDDQRTEMPRPKADPRPKTVPDDDFFALIMRAQSGRMEDQRAAIPKPAENRQSKPAPKPRK
Summary
Description
S-adenosyl-L-methionine-binding protein that acts as an inhibitor of mTORC1 signaling. Acts as a sensor of S-adenosyl-L-methionine to signal methionine sufficiency to mTORC1. Probably also acts as a S-adenosyl-L-methionine-dependent methyltransferase.
Similarity
Belongs to the BMT2 family.
Uniprot
A0A1E1W3B1
H9J2Y0
A0A2H1WJV0
A0A2A4JQ38
A0A194QXQ7
A0A194PNK0
+ More
A0A2P8XTP3 E0VT28 T1HV42 A0A067QMN4 A0A023F5F5 A0A2J7RIX7 A0A0P4VIV9 A0A069DV90 A0A212EKE1 N6THL1 A0A2H8TGF8 D6WEG2 A0A0A9YDJ9 A0A1W4XHA3 J9JZ96 A0A1S4EGW1 A0A2J7RIZ1 A0A1Y1JRY1 A0A3Q0J238 A0A1W4XRU7 A0A0A9YJN0 A0A1Q3FAH9 A0A1Q3FAP2 A0A1Q3FAL2 A0A0P6IVL0 A0A1Q3FAP3 A0A2S2PZV3 A0A1S4FKG9 B0WSA6 E9FQZ4 A0A1L8DLR2 A0A0P6AQT7 Q7PWG1 A0A3Q0J269 A0A1Y1JV02 A0A0P5W7S2 A0A0M9A5N8 A0A088A1H2 E2C2Q2 A0A026WI13 A0A154PMH8 A0A0L7QNN5 A0A182PVU5 A0A182M4F9 K7IUG1 A0A0K8TCG7 A0A182Y0H7 A0A195DTF7 A0A182RZB7 A0A164U0Q6 A0A2A3EJP5 A0A0C9QFA0 A0A195BLK8 Q16Y24 A0A195CCN6 A0A0J7L357 F4X2Q8 A0A195FVU9 Q16H10 A0A3L8DIJ5 E9J298 A0A182LMJ7 A0A182I7I7 A0A1J1JA02 B4QXH8 A0A182Q4N9 B4IGT6 A0A1S4H0I0 A0A1B6CEP6 A0A1B6E487 Q29BH6 B4GNY6 A0A182WXM2 A0A2M4BES0 B3LXJ2 B4JRY6 A0A2M4BEL2 B4M633 A0A0N7ZLK7 A0A0M3QXH9 A0A0K8VPI6 W8C4I9 A0A1B6EXM6 A0A2J7RIY8 A0A182FN86 A0A182JI38 A0A0K8TCW5 A0A182WGC1 A0A0A1WVH3 A0A0P4W3L7 A0A210QBM3 A0A182V7B0 A0A182NBJ6 A0A1S3KCE9
A0A2P8XTP3 E0VT28 T1HV42 A0A067QMN4 A0A023F5F5 A0A2J7RIX7 A0A0P4VIV9 A0A069DV90 A0A212EKE1 N6THL1 A0A2H8TGF8 D6WEG2 A0A0A9YDJ9 A0A1W4XHA3 J9JZ96 A0A1S4EGW1 A0A2J7RIZ1 A0A1Y1JRY1 A0A3Q0J238 A0A1W4XRU7 A0A0A9YJN0 A0A1Q3FAH9 A0A1Q3FAP2 A0A1Q3FAL2 A0A0P6IVL0 A0A1Q3FAP3 A0A2S2PZV3 A0A1S4FKG9 B0WSA6 E9FQZ4 A0A1L8DLR2 A0A0P6AQT7 Q7PWG1 A0A3Q0J269 A0A1Y1JV02 A0A0P5W7S2 A0A0M9A5N8 A0A088A1H2 E2C2Q2 A0A026WI13 A0A154PMH8 A0A0L7QNN5 A0A182PVU5 A0A182M4F9 K7IUG1 A0A0K8TCG7 A0A182Y0H7 A0A195DTF7 A0A182RZB7 A0A164U0Q6 A0A2A3EJP5 A0A0C9QFA0 A0A195BLK8 Q16Y24 A0A195CCN6 A0A0J7L357 F4X2Q8 A0A195FVU9 Q16H10 A0A3L8DIJ5 E9J298 A0A182LMJ7 A0A182I7I7 A0A1J1JA02 B4QXH8 A0A182Q4N9 B4IGT6 A0A1S4H0I0 A0A1B6CEP6 A0A1B6E487 Q29BH6 B4GNY6 A0A182WXM2 A0A2M4BES0 B3LXJ2 B4JRY6 A0A2M4BEL2 B4M633 A0A0N7ZLK7 A0A0M3QXH9 A0A0K8VPI6 W8C4I9 A0A1B6EXM6 A0A2J7RIY8 A0A182FN86 A0A182JI38 A0A0K8TCW5 A0A182WGC1 A0A0A1WVH3 A0A0P4W3L7 A0A210QBM3 A0A182V7B0 A0A182NBJ6 A0A1S3KCE9
Pubmed
EMBL
GDQN01009586
JAT81468.1
BABH01031212
ODYU01009120
SOQ53318.1
NWSH01000836
+ More
PCG73929.1 KQ460953 KPJ10323.1 KQ459599 KPI94319.1 PYGN01001366 PSN35366.1 DS235759 EEB16534.1 ACPB03018893 KK853156 KDR10512.1 GBBI01002338 JAC16374.1 NEVH01003011 PNF40798.1 GDKW01001754 JAI54841.1 GBGD01000906 JAC87983.1 AGBW02014274 OWR41951.1 APGK01026647 KB740635 KB632195 ENN79909.1 ERL89968.1 GFXV01001392 MBW13197.1 KQ971318 EFA00406.2 GBHO01013890 GBHO01013888 GBHO01013886 GBRD01002571 GDHC01012135 JAG29714.1 JAG29716.1 JAG29718.1 JAG63250.1 JAQ06494.1 ABLF02009781 PNF40797.1 GEZM01102118 JAV51952.1 GBHO01013889 GBHO01013887 GBHO01013885 GBRD01002572 JAG29715.1 JAG29717.1 JAG29719.1 JAG63249.1 GFDL01010485 JAV24560.1 GFDL01010463 JAV24582.1 GFDL01010420 JAV24625.1 GDUN01000717 JAN95202.1 GFDL01010456 JAV24589.1 GGMS01001229 MBY70432.1 DS232067 EDS33736.1 GL732523 EFX90280.1 GFDF01006777 JAV07307.1 GDIP01027515 JAM76200.1 AAAB01008984 EAA14880.4 GEZM01102119 JAV51951.1 GDIP01090097 JAM13618.1 KQ435748 KOX76463.1 GL452203 EFN77737.1 KK107238 EZA54744.1 KQ434978 KZC13043.1 KQ414851 KOC60235.1 AXCM01006472 AAZX01008038 GBRD01002573 JAG63248.1 KQ980489 KYN15824.1 LRGB01001654 KZS10949.1 KZ288241 PBC31261.1 GBYB01002074 JAG71841.1 KQ976450 KYM85687.1 CH477527 EAT39510.1 KQ977957 KYM98475.1 LBMM01001122 KMQ96884.1 GL888591 EGI59254.1 KQ981219 KYN44432.1 CH478216 EAT33528.1 QOIP01000007 RLU20237.1 GL767834 EFZ13039.1 APCN01003409 CVRI01000075 CRL08700.1 CM000364 EDX14664.1 AXCN02000674 CH480837 EDW49054.1 GEDC01025385 JAS11913.1 GEDC01004567 JAS32731.1 CM000070 EAL27022.1 CH479186 EDW38869.1 GGFJ01002352 MBW51493.1 CH902617 EDV43886.1 CH916373 EDV94526.1 GGFJ01002344 MBW51485.1 CH940652 EDW59109.1 GDIP01233481 JAI89920.1 CP012526 ALC45953.1 GDHF01031339 GDHF01025535 GDHF01023549 GDHF01011520 JAI20975.1 JAI26779.1 JAI28765.1 JAI40794.1 GAMC01005479 JAC01077.1 GECZ01027139 JAS42630.1 PNF40799.1 GBRD01002570 JAG63251.1 GBXI01011762 JAD02530.1 GDRN01090458 JAI60468.1 NEDP02004270 OWF46128.1
PCG73929.1 KQ460953 KPJ10323.1 KQ459599 KPI94319.1 PYGN01001366 PSN35366.1 DS235759 EEB16534.1 ACPB03018893 KK853156 KDR10512.1 GBBI01002338 JAC16374.1 NEVH01003011 PNF40798.1 GDKW01001754 JAI54841.1 GBGD01000906 JAC87983.1 AGBW02014274 OWR41951.1 APGK01026647 KB740635 KB632195 ENN79909.1 ERL89968.1 GFXV01001392 MBW13197.1 KQ971318 EFA00406.2 GBHO01013890 GBHO01013888 GBHO01013886 GBRD01002571 GDHC01012135 JAG29714.1 JAG29716.1 JAG29718.1 JAG63250.1 JAQ06494.1 ABLF02009781 PNF40797.1 GEZM01102118 JAV51952.1 GBHO01013889 GBHO01013887 GBHO01013885 GBRD01002572 JAG29715.1 JAG29717.1 JAG29719.1 JAG63249.1 GFDL01010485 JAV24560.1 GFDL01010463 JAV24582.1 GFDL01010420 JAV24625.1 GDUN01000717 JAN95202.1 GFDL01010456 JAV24589.1 GGMS01001229 MBY70432.1 DS232067 EDS33736.1 GL732523 EFX90280.1 GFDF01006777 JAV07307.1 GDIP01027515 JAM76200.1 AAAB01008984 EAA14880.4 GEZM01102119 JAV51951.1 GDIP01090097 JAM13618.1 KQ435748 KOX76463.1 GL452203 EFN77737.1 KK107238 EZA54744.1 KQ434978 KZC13043.1 KQ414851 KOC60235.1 AXCM01006472 AAZX01008038 GBRD01002573 JAG63248.1 KQ980489 KYN15824.1 LRGB01001654 KZS10949.1 KZ288241 PBC31261.1 GBYB01002074 JAG71841.1 KQ976450 KYM85687.1 CH477527 EAT39510.1 KQ977957 KYM98475.1 LBMM01001122 KMQ96884.1 GL888591 EGI59254.1 KQ981219 KYN44432.1 CH478216 EAT33528.1 QOIP01000007 RLU20237.1 GL767834 EFZ13039.1 APCN01003409 CVRI01000075 CRL08700.1 CM000364 EDX14664.1 AXCN02000674 CH480837 EDW49054.1 GEDC01025385 JAS11913.1 GEDC01004567 JAS32731.1 CM000070 EAL27022.1 CH479186 EDW38869.1 GGFJ01002352 MBW51493.1 CH902617 EDV43886.1 CH916373 EDV94526.1 GGFJ01002344 MBW51485.1 CH940652 EDW59109.1 GDIP01233481 JAI89920.1 CP012526 ALC45953.1 GDHF01031339 GDHF01025535 GDHF01023549 GDHF01011520 JAI20975.1 JAI26779.1 JAI28765.1 JAI40794.1 GAMC01005479 JAC01077.1 GECZ01027139 JAS42630.1 PNF40799.1 GBRD01002570 JAG63251.1 GBXI01011762 JAD02530.1 GDRN01090458 JAI60468.1 NEDP02004270 OWF46128.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000245037
UP000009046
+ More
UP000015103 UP000027135 UP000235965 UP000007151 UP000019118 UP000030742 UP000007266 UP000192223 UP000007819 UP000079169 UP000002320 UP000000305 UP000007062 UP000053105 UP000005203 UP000008237 UP000053097 UP000076502 UP000053825 UP000075885 UP000075883 UP000002358 UP000076408 UP000078492 UP000075900 UP000076858 UP000242457 UP000078540 UP000008820 UP000078542 UP000036403 UP000007755 UP000078541 UP000279307 UP000075882 UP000075840 UP000183832 UP000000304 UP000075886 UP000001292 UP000001819 UP000008744 UP000076407 UP000007801 UP000001070 UP000008792 UP000092553 UP000069272 UP000075880 UP000075920 UP000242188 UP000075903 UP000075884 UP000085678
UP000015103 UP000027135 UP000235965 UP000007151 UP000019118 UP000030742 UP000007266 UP000192223 UP000007819 UP000079169 UP000002320 UP000000305 UP000007062 UP000053105 UP000005203 UP000008237 UP000053097 UP000076502 UP000053825 UP000075885 UP000075883 UP000002358 UP000076408 UP000078492 UP000075900 UP000076858 UP000242457 UP000078540 UP000008820 UP000078542 UP000036403 UP000007755 UP000078541 UP000279307 UP000075882 UP000075840 UP000183832 UP000000304 UP000075886 UP000001292 UP000001819 UP000008744 UP000076407 UP000007801 UP000001070 UP000008792 UP000092553 UP000069272 UP000075880 UP000075920 UP000242188 UP000075903 UP000075884 UP000085678
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A1E1W3B1
H9J2Y0
A0A2H1WJV0
A0A2A4JQ38
A0A194QXQ7
A0A194PNK0
+ More
A0A2P8XTP3 E0VT28 T1HV42 A0A067QMN4 A0A023F5F5 A0A2J7RIX7 A0A0P4VIV9 A0A069DV90 A0A212EKE1 N6THL1 A0A2H8TGF8 D6WEG2 A0A0A9YDJ9 A0A1W4XHA3 J9JZ96 A0A1S4EGW1 A0A2J7RIZ1 A0A1Y1JRY1 A0A3Q0J238 A0A1W4XRU7 A0A0A9YJN0 A0A1Q3FAH9 A0A1Q3FAP2 A0A1Q3FAL2 A0A0P6IVL0 A0A1Q3FAP3 A0A2S2PZV3 A0A1S4FKG9 B0WSA6 E9FQZ4 A0A1L8DLR2 A0A0P6AQT7 Q7PWG1 A0A3Q0J269 A0A1Y1JV02 A0A0P5W7S2 A0A0M9A5N8 A0A088A1H2 E2C2Q2 A0A026WI13 A0A154PMH8 A0A0L7QNN5 A0A182PVU5 A0A182M4F9 K7IUG1 A0A0K8TCG7 A0A182Y0H7 A0A195DTF7 A0A182RZB7 A0A164U0Q6 A0A2A3EJP5 A0A0C9QFA0 A0A195BLK8 Q16Y24 A0A195CCN6 A0A0J7L357 F4X2Q8 A0A195FVU9 Q16H10 A0A3L8DIJ5 E9J298 A0A182LMJ7 A0A182I7I7 A0A1J1JA02 B4QXH8 A0A182Q4N9 B4IGT6 A0A1S4H0I0 A0A1B6CEP6 A0A1B6E487 Q29BH6 B4GNY6 A0A182WXM2 A0A2M4BES0 B3LXJ2 B4JRY6 A0A2M4BEL2 B4M633 A0A0N7ZLK7 A0A0M3QXH9 A0A0K8VPI6 W8C4I9 A0A1B6EXM6 A0A2J7RIY8 A0A182FN86 A0A182JI38 A0A0K8TCW5 A0A182WGC1 A0A0A1WVH3 A0A0P4W3L7 A0A210QBM3 A0A182V7B0 A0A182NBJ6 A0A1S3KCE9
A0A2P8XTP3 E0VT28 T1HV42 A0A067QMN4 A0A023F5F5 A0A2J7RIX7 A0A0P4VIV9 A0A069DV90 A0A212EKE1 N6THL1 A0A2H8TGF8 D6WEG2 A0A0A9YDJ9 A0A1W4XHA3 J9JZ96 A0A1S4EGW1 A0A2J7RIZ1 A0A1Y1JRY1 A0A3Q0J238 A0A1W4XRU7 A0A0A9YJN0 A0A1Q3FAH9 A0A1Q3FAP2 A0A1Q3FAL2 A0A0P6IVL0 A0A1Q3FAP3 A0A2S2PZV3 A0A1S4FKG9 B0WSA6 E9FQZ4 A0A1L8DLR2 A0A0P6AQT7 Q7PWG1 A0A3Q0J269 A0A1Y1JV02 A0A0P5W7S2 A0A0M9A5N8 A0A088A1H2 E2C2Q2 A0A026WI13 A0A154PMH8 A0A0L7QNN5 A0A182PVU5 A0A182M4F9 K7IUG1 A0A0K8TCG7 A0A182Y0H7 A0A195DTF7 A0A182RZB7 A0A164U0Q6 A0A2A3EJP5 A0A0C9QFA0 A0A195BLK8 Q16Y24 A0A195CCN6 A0A0J7L357 F4X2Q8 A0A195FVU9 Q16H10 A0A3L8DIJ5 E9J298 A0A182LMJ7 A0A182I7I7 A0A1J1JA02 B4QXH8 A0A182Q4N9 B4IGT6 A0A1S4H0I0 A0A1B6CEP6 A0A1B6E487 Q29BH6 B4GNY6 A0A182WXM2 A0A2M4BES0 B3LXJ2 B4JRY6 A0A2M4BEL2 B4M633 A0A0N7ZLK7 A0A0M3QXH9 A0A0K8VPI6 W8C4I9 A0A1B6EXM6 A0A2J7RIY8 A0A182FN86 A0A182JI38 A0A0K8TCW5 A0A182WGC1 A0A0A1WVH3 A0A0P4W3L7 A0A210QBM3 A0A182V7B0 A0A182NBJ6 A0A1S3KCE9
PDB
5A7D
E-value=2.08936e-140,
Score=1281
Ontologies
GO
Topology
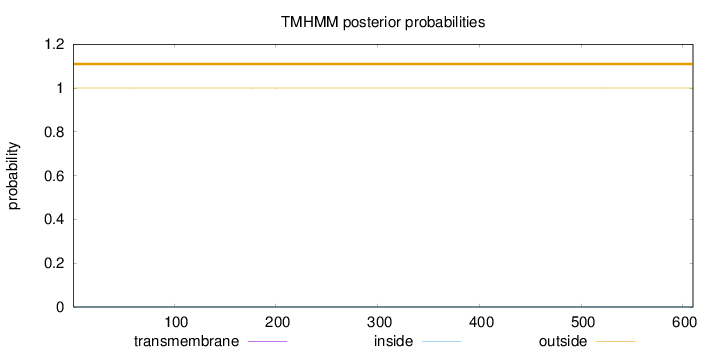
Length:
610
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00408
Exp number, first 60 AAs:
0.00211
Total prob of N-in:
0.00025
outside
1 - 610
Population Genetic Test Statistics
Pi
22.898133
Theta
18.286145
Tajima's D
-1.329349
CLR
0.075213
CSRT
0.0834458277086146
Interpretation
Uncertain
