Gene
KWMTBOMO00649
Pre Gene Modal
BGIBMGA003889
Annotation
PREDICTED:_ATP_synthase_subunit_gamma?_mitochondrial-like_[Amyelois_transitella]
Full name
ATP synthase subunit gamma
Location in the cell
Mitochondrial Reliability : 2.425
Sequence
CDS
ATGGTACAGTTGAAACAAGTGTCCCTAAGATTAAAATCGATAAAAAACATACAGAAGATTACCAAAACTATGAAAATGGTTTCCGCTAGCAAGTTTACAAAAGCAGAACGCGAACTGCAAGCAGCGAGGCCTTTCGGTTATGGCCCTAGGAAGTTCTACGAATCTTCTCGATTGATAAAGGCGGCTGTGGACAGCAAAGATAAGAAAGACGCCGCCGCTGTCCCCGAACCAGTGCAGGAGAAACCGGAAGACCAGGAGGCCCAAAAGAAACTCAAAAGGATTTATATAGCTATGACGTCTGACAGAGGTATCTAA
Protein
MVQLKQVSLRLKSIKNIQKITKTMKMVSASKFTKAERELQAARPFGYGPRKFYESSRLIKAAVDSKDKKDAAAVPEPVQEKPEDQEAQKKLKRIYIAMTSDRGI
Summary
Subunit
F-type ATPases have 2 components, CF(1) - the catalytic core - and CF(0) - the membrane proton channel. CF(1) has five subunits: alpha(3), beta(3), gamma(1), delta(1), epsilon(1). CF(0) has three main subunits: a, b and c.
Similarity
Belongs to the ATPase gamma chain family.
Feature
chain ATP synthase subunit gamma
Uniprot
A0A212EKD8
A0A2W1BPS2
A0A2H1VKF2
A0A2A4JR83
H9J302
A0A194PLV8
+ More
A0A194QZB7 G9D524 G9D525 G9D521 G9D529 G9D528 G9D531 G9D522 G9D520 G9D527 G9D530 G9D523 G9D519 G9D526 G9D518 A0A3G1T1C7 A0A2A4JBB2 A0A1I8QC49 A0A095A1I0 A0A183M325 A0A183KX31 I4DJ96 S4NNE5 A0A212F7F8 T1PF90 A0A2W1B7N2 A0A2H1WBL5 A0A183WF96 A0A3P8JAL4 W8BR16 A0A0N4Z0G2 A0A1A9X5G8 A0A1Y1LVM0 A0A0K0FN58 G4LVB6 A0A182JWN3 A0A2P2HWM3 A0A1L8EEQ6 A0A0L0C3Z3 G4LVB9 A0A3B0EA27 Q1HPX4 T1JKD0 A0A183QA92 A0A0A1WI89 A0A182NTJ6 A0A182PD01 A0A182QIX0 A0A194QVY4 Q7Q3N8 A0A182XZU4 A0A182UP01 A0A182L1Z8 A0A182UFE6 A0A182HV43 A0A182LYU4 A0A182W7I1 A0A182WT48 Q5DBP4 A0A1D2N7T7 E0VFT5 C1LBB8 A0A1A9VLY0 D3TMX6 A0A0K0EQT5 A0A1W4WJB5 A0A310SJI5 A0A1B0AFH2 B4G663 Q29AN8 A0A210Q163 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 A0A0A9Z7I7 B0FWR7 A0A0K8U3G5 A0A0T6BGI0 A0A1B0FAP1 A0A182RJB1 C1LVF7 A0A1B6MUF0 A0A2R7WX91 A0A2M3YZ82 A0A023ENY5 A3EY11 Q16XK3 W5JNX9 A0A0N5BRZ9 A0A2M4A2J6 A0A182F302 B3MTI4 A0A0M4ET73
A0A194QZB7 G9D524 G9D525 G9D521 G9D529 G9D528 G9D531 G9D522 G9D520 G9D527 G9D530 G9D523 G9D519 G9D526 G9D518 A0A3G1T1C7 A0A2A4JBB2 A0A1I8QC49 A0A095A1I0 A0A183M325 A0A183KX31 I4DJ96 S4NNE5 A0A212F7F8 T1PF90 A0A2W1B7N2 A0A2H1WBL5 A0A183WF96 A0A3P8JAL4 W8BR16 A0A0N4Z0G2 A0A1A9X5G8 A0A1Y1LVM0 A0A0K0FN58 G4LVB6 A0A182JWN3 A0A2P2HWM3 A0A1L8EEQ6 A0A0L0C3Z3 G4LVB9 A0A3B0EA27 Q1HPX4 T1JKD0 A0A183QA92 A0A0A1WI89 A0A182NTJ6 A0A182PD01 A0A182QIX0 A0A194QVY4 Q7Q3N8 A0A182XZU4 A0A182UP01 A0A182L1Z8 A0A182UFE6 A0A182HV43 A0A182LYU4 A0A182W7I1 A0A182WT48 Q5DBP4 A0A1D2N7T7 E0VFT5 C1LBB8 A0A1A9VLY0 D3TMX6 A0A0K0EQT5 A0A1W4WJB5 A0A310SJI5 A0A1B0AFH2 B4G663 Q29AN8 A0A210Q163 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 A0A0A9Z7I7 B0FWR7 A0A0K8U3G5 A0A0T6BGI0 A0A1B0FAP1 A0A182RJB1 C1LVF7 A0A1B6MUF0 A0A2R7WX91 A0A2M3YZ82 A0A023ENY5 A3EY11 Q16XK3 W5JNX9 A0A0N5BRZ9 A0A2M4A2J6 A0A182F302 B3MTI4 A0A0M4ET73
Pubmed
22118469
28756777
19121390
26354079
21967427
22246508
+ More
22651552 23622113 25315136 24495485 28004739 22253936 26108605 25830018 12364791 14747013 17210077 25244985 20966253 16617374 27289101 20566863 20353571 17994087 15632085 23185243 28812685 25348373 25401762 26823975 24945155 26483478 17510324 20920257 23761445
22651552 23622113 25315136 24495485 28004739 22253936 26108605 25830018 12364791 14747013 17210077 25244985 20966253 16617374 27289101 20566863 20353571 17994087 15632085 23185243 28812685 25348373 25401762 26823975 24945155 26483478 17510324 20920257 23761445
EMBL
AGBW02014274
OWR41946.1
KZ150031
PZC74730.1
ODYU01003024
SOQ41271.1
+ More
NWSH01000836 PCG73932.1 BABH01031222 BABH01031223 KQ459599 KPI94312.1 KQ460953 KPJ10320.1 JF777427 AEU11772.1 JF777428 AEU11773.1 JF777424 AEU11769.1 JF777432 AEU11777.1 JF777431 AEU11776.1 JF777434 AEU11779.1 JF777425 AEU11770.1 JF777423 AEU11768.1 JF777430 AEU11775.1 JF777433 AEU11778.1 JF777426 AEU11771.1 JF777422 AEU11767.1 JF777429 AEU11774.1 JF777421 AEU11766.1 MG846921 AXY94773.1 NWSH01002112 PCG69139.1 KL251729 KGB41130.1 UZAI01005408 VDO90413.1 UZAK01042841 VDP69724.1 AK401364 KQ459583 BAM17986.1 KPI98617.1 GAIX01012289 JAA80271.1 AGBW02009877 OWR49666.1 KA647354 AFP61983.1 KZ150244 PZC71889.1 ODYU01007571 SOQ50453.1 UZAO01067245 VDQ06679.1 GAMC01010884 JAB95671.1 GEZM01050230 GEZM01050229 JAV75666.1 CABG01000013 CCD58855.1 IACF01000394 LAB66171.1 GFDG01001618 JAV17181.1 JRES01001003 KNC26199.1 CCD58858.1 RBVL01000238 RKO10274.1 BABH01018421 DQ443278 ABF51367.1 JH431830 GBXI01015931 JAC98360.1 AXCN02000699 KQ461103 KPJ09474.1 AAAB01008964 EAA12854.3 APCN01002493 AXCM01002571 AY815030 FN316264 FN316266 FN316267 FN316268 FN316269 FN316270 FN316271 FN316272 FN316273 FN322956 FN322959 FN322960 FN322962 FN322963 FN322964 FN322965 FN322966 FN322967 FN322968 FN322969 FN322970 FN322971 FN322972 AAW26762.1 CAX71995.1 LJIJ01000160 ODN01324.1 DS235124 EEB12241.1 FN316265 CAX71996.1 EZ422778 ADD19054.1 KQ760551 OAD59818.1 CH479179 EDW23816.1 CM000070 EAL27311.1 NEDP02005265 OWF42490.1 GDHF01012143 JAI40171.1 GAKP01006469 JAC52483.1 GBYB01006053 JAG75820.1 GBHO01004281 GDHC01013990 JAG39323.1 JAQ04639.1 EU358107 ABY62749.1 GDHF01031075 JAI21239.1 LJIG01000515 KRT86425.1 CCAG010009218 FN322961 CAX78685.1 GEBQ01000422 JAT39555.1 KK855861 PTY24152.1 GGFM01000831 MBW21582.1 JXUM01116988 GAPW01002938 KQ566040 JAC10660.1 KXJ70434.1 EF092079 ABN12071.1 CH477537 EAT39355.1 ADMH02000571 ETN65841.1 GGFK01001696 MBW35017.1 CH902623 EDV30574.1 CP012526 ALC48060.1
NWSH01000836 PCG73932.1 BABH01031222 BABH01031223 KQ459599 KPI94312.1 KQ460953 KPJ10320.1 JF777427 AEU11772.1 JF777428 AEU11773.1 JF777424 AEU11769.1 JF777432 AEU11777.1 JF777431 AEU11776.1 JF777434 AEU11779.1 JF777425 AEU11770.1 JF777423 AEU11768.1 JF777430 AEU11775.1 JF777433 AEU11778.1 JF777426 AEU11771.1 JF777422 AEU11767.1 JF777429 AEU11774.1 JF777421 AEU11766.1 MG846921 AXY94773.1 NWSH01002112 PCG69139.1 KL251729 KGB41130.1 UZAI01005408 VDO90413.1 UZAK01042841 VDP69724.1 AK401364 KQ459583 BAM17986.1 KPI98617.1 GAIX01012289 JAA80271.1 AGBW02009877 OWR49666.1 KA647354 AFP61983.1 KZ150244 PZC71889.1 ODYU01007571 SOQ50453.1 UZAO01067245 VDQ06679.1 GAMC01010884 JAB95671.1 GEZM01050230 GEZM01050229 JAV75666.1 CABG01000013 CCD58855.1 IACF01000394 LAB66171.1 GFDG01001618 JAV17181.1 JRES01001003 KNC26199.1 CCD58858.1 RBVL01000238 RKO10274.1 BABH01018421 DQ443278 ABF51367.1 JH431830 GBXI01015931 JAC98360.1 AXCN02000699 KQ461103 KPJ09474.1 AAAB01008964 EAA12854.3 APCN01002493 AXCM01002571 AY815030 FN316264 FN316266 FN316267 FN316268 FN316269 FN316270 FN316271 FN316272 FN316273 FN322956 FN322959 FN322960 FN322962 FN322963 FN322964 FN322965 FN322966 FN322967 FN322968 FN322969 FN322970 FN322971 FN322972 AAW26762.1 CAX71995.1 LJIJ01000160 ODN01324.1 DS235124 EEB12241.1 FN316265 CAX71996.1 EZ422778 ADD19054.1 KQ760551 OAD59818.1 CH479179 EDW23816.1 CM000070 EAL27311.1 NEDP02005265 OWF42490.1 GDHF01012143 JAI40171.1 GAKP01006469 JAC52483.1 GBYB01006053 JAG75820.1 GBHO01004281 GDHC01013990 JAG39323.1 JAQ04639.1 EU358107 ABY62749.1 GDHF01031075 JAI21239.1 LJIG01000515 KRT86425.1 CCAG010009218 FN322961 CAX78685.1 GEBQ01000422 JAT39555.1 KK855861 PTY24152.1 GGFM01000831 MBW21582.1 JXUM01116988 GAPW01002938 KQ566040 JAC10660.1 KXJ70434.1 EF092079 ABN12071.1 CH477537 EAT39355.1 ADMH02000571 ETN65841.1 GGFK01001696 MBW35017.1 CH902623 EDV30574.1 CP012526 ALC48060.1
Proteomes
UP000007151
UP000218220
UP000005204
UP000053268
UP000053240
UP000095300
+ More
UP000050790 UP000277204 UP000050789 UP000095301 UP000050795 UP000038045 UP000091820 UP000035680 UP000008854 UP000075881 UP000037069 UP000276569 UP000050792 UP000075884 UP000075885 UP000075886 UP000007062 UP000076408 UP000075903 UP000075882 UP000075902 UP000075840 UP000075883 UP000075920 UP000076407 UP000094527 UP000009046 UP000078200 UP000035681 UP000192223 UP000092445 UP000008744 UP000001819 UP000242188 UP000092444 UP000075900 UP000069940 UP000249989 UP000008820 UP000000673 UP000046392 UP000069272 UP000007801 UP000092553
UP000050790 UP000277204 UP000050789 UP000095301 UP000050795 UP000038045 UP000091820 UP000035680 UP000008854 UP000075881 UP000037069 UP000276569 UP000050792 UP000075884 UP000075885 UP000075886 UP000007062 UP000076408 UP000075903 UP000075882 UP000075902 UP000075840 UP000075883 UP000075920 UP000076407 UP000094527 UP000009046 UP000078200 UP000035681 UP000192223 UP000092445 UP000008744 UP000001819 UP000242188 UP000092444 UP000075900 UP000069940 UP000249989 UP000008820 UP000000673 UP000046392 UP000069272 UP000007801 UP000092553
Interpro
Gene 3D
ProteinModelPortal
A0A212EKD8
A0A2W1BPS2
A0A2H1VKF2
A0A2A4JR83
H9J302
A0A194PLV8
+ More
A0A194QZB7 G9D524 G9D525 G9D521 G9D529 G9D528 G9D531 G9D522 G9D520 G9D527 G9D530 G9D523 G9D519 G9D526 G9D518 A0A3G1T1C7 A0A2A4JBB2 A0A1I8QC49 A0A095A1I0 A0A183M325 A0A183KX31 I4DJ96 S4NNE5 A0A212F7F8 T1PF90 A0A2W1B7N2 A0A2H1WBL5 A0A183WF96 A0A3P8JAL4 W8BR16 A0A0N4Z0G2 A0A1A9X5G8 A0A1Y1LVM0 A0A0K0FN58 G4LVB6 A0A182JWN3 A0A2P2HWM3 A0A1L8EEQ6 A0A0L0C3Z3 G4LVB9 A0A3B0EA27 Q1HPX4 T1JKD0 A0A183QA92 A0A0A1WI89 A0A182NTJ6 A0A182PD01 A0A182QIX0 A0A194QVY4 Q7Q3N8 A0A182XZU4 A0A182UP01 A0A182L1Z8 A0A182UFE6 A0A182HV43 A0A182LYU4 A0A182W7I1 A0A182WT48 Q5DBP4 A0A1D2N7T7 E0VFT5 C1LBB8 A0A1A9VLY0 D3TMX6 A0A0K0EQT5 A0A1W4WJB5 A0A310SJI5 A0A1B0AFH2 B4G663 Q29AN8 A0A210Q163 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 A0A0A9Z7I7 B0FWR7 A0A0K8U3G5 A0A0T6BGI0 A0A1B0FAP1 A0A182RJB1 C1LVF7 A0A1B6MUF0 A0A2R7WX91 A0A2M3YZ82 A0A023ENY5 A3EY11 Q16XK3 W5JNX9 A0A0N5BRZ9 A0A2M4A2J6 A0A182F302 B3MTI4 A0A0M4ET73
A0A194QZB7 G9D524 G9D525 G9D521 G9D529 G9D528 G9D531 G9D522 G9D520 G9D527 G9D530 G9D523 G9D519 G9D526 G9D518 A0A3G1T1C7 A0A2A4JBB2 A0A1I8QC49 A0A095A1I0 A0A183M325 A0A183KX31 I4DJ96 S4NNE5 A0A212F7F8 T1PF90 A0A2W1B7N2 A0A2H1WBL5 A0A183WF96 A0A3P8JAL4 W8BR16 A0A0N4Z0G2 A0A1A9X5G8 A0A1Y1LVM0 A0A0K0FN58 G4LVB6 A0A182JWN3 A0A2P2HWM3 A0A1L8EEQ6 A0A0L0C3Z3 G4LVB9 A0A3B0EA27 Q1HPX4 T1JKD0 A0A183QA92 A0A0A1WI89 A0A182NTJ6 A0A182PD01 A0A182QIX0 A0A194QVY4 Q7Q3N8 A0A182XZU4 A0A182UP01 A0A182L1Z8 A0A182UFE6 A0A182HV43 A0A182LYU4 A0A182W7I1 A0A182WT48 Q5DBP4 A0A1D2N7T7 E0VFT5 C1LBB8 A0A1A9VLY0 D3TMX6 A0A0K0EQT5 A0A1W4WJB5 A0A310SJI5 A0A1B0AFH2 B4G663 Q29AN8 A0A210Q163 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 A0A0A9Z7I7 B0FWR7 A0A0K8U3G5 A0A0T6BGI0 A0A1B0FAP1 A0A182RJB1 C1LVF7 A0A1B6MUF0 A0A2R7WX91 A0A2M3YZ82 A0A023ENY5 A3EY11 Q16XK3 W5JNX9 A0A0N5BRZ9 A0A2M4A2J6 A0A182F302 B3MTI4 A0A0M4ET73
PDB
2W6J
E-value=1.89969e-11,
Score=160
Ontologies
PATHWAY
GO
PANTHER
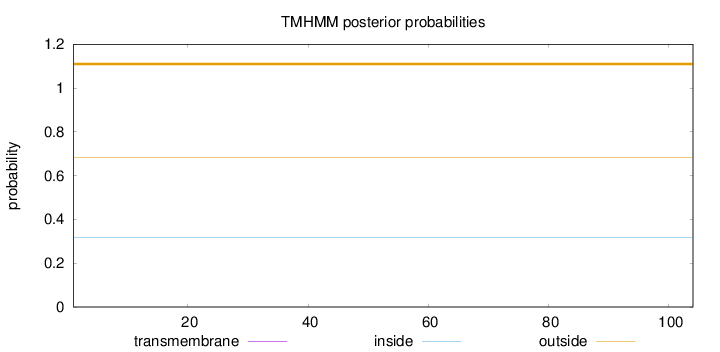
Topology
Length:
104
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.31656
outside
1 - 104
Population Genetic Test Statistics
Pi
11.0478
Theta
16.392153
Tajima's D
-1.200388
CLR
0
CSRT
0.101994900254987
Interpretation
Uncertain
