Gene
KWMTBOMO00648
Pre Gene Modal
BGIBMGA003889
Annotation
ATP_synthase_subunit_gamma_[Operophtera_brumata]
Full name
ATP synthase subunit gamma
+ More
ATP synthase subunit gamma, mitochondrial
ATP synthase subunit gamma, mitochondrial
Alternative Name
F-ATPase gamma subunit
Location in the cell
Mitochondrial Reliability : 1.592
Sequence
CDS
ATGACCGCTAGGAACGCCGAGGGCACGACACACAAGCTGGTGTGCGTCGGCGACAAGGCTCGGGCGTTGCTGCGCCGGGCGTACTCCACCAGCATGCTGGTCACCGTTAAAGATATCGGTCGCATAGCCCCTGTGTTCGCCGACGCGTCACTTATGGCAACCGCGATCGCCGAGTCGGGCTTCACTTACGACGTCGGTGAGATCTACTACAACAAGTACTTTACGGCGGTCAAATATGAAATGTCCATTGTGCCATTCTTCAGTAAAATCAGGATTGAGGTAGGTTGTATCACACAAATATACGTATCACACAAGACATCTCGATAG
Protein
MTARNAEGTTHKLVCVGDKARALLRRAYSTSMLVTVKDIGRIAPVFADASLMATAIAESGFTYDVGEIYYNKYFTAVKYEMSIVPFFSKIRIEVGCITQIYVSHKTSR
Summary
Subunit
F-type ATPases have 2 components, CF(1) - the catalytic core - and CF(0) - the membrane proton channel. CF(1) has five subunits: alpha(3), beta(3), gamma(1), delta(1), epsilon(1). CF(0) has three main subunits: a, b and c.
Similarity
Belongs to the ATPase gamma chain family.
Keywords
ATP synthesis
Complete proteome
Hydrogen ion transport
Ion transport
Membrane
Mitochondrion
Mitochondrion inner membrane
Reference proteome
Transit peptide
Transport
Feature
chain ATP synthase subunit gamma, mitochondrial
Uniprot
H9J302
A0A0L7LA84
A0A2H1VKF2
A0A2A4JR52
A0A2A4JR83
A0A2W1BPS2
+ More
A0A212EKD8 A0A194PLV8 A0A194QZB7 A0A1Y1LVM0 A0A1I8C016 A0A0V1JZ94 A0A0V0YCW8 A0A0V1NT42 A0A1Y3EIA8 A0A0V0VAI0 A0A0V0WD05 A0A0V0UDS0 A0A0V1A3F7 A0A0V1CQD4 A0A1D2N7T7 A0A267G1M7 A0A1I8HV25 E5RZD5 A0A0V1EVZ2 A0A1I8H306 A0A0V1L6C9 A0A0V1BZ30 C1BPC0 A0A210Q163 A0A0V1MFY0 A0A0U2M8V4 A0A0V1GYS8 A0A158PEG7 A0A3G1T1C7 A0A183FFX2 A0A3P7Y210 G9D519 A0A2H1WBL5 G9D526 S4NNE5 A0A077ZAI2 Q1HPX4 A0A0N4XXV6 A0A1D1UVH4 B6V839 R7UAF7 A0A085MJX5 A0A0N5E5B1 A0A0K2T9M4 A0A3M7SU40 C1BTJ1 A0A1V9XES2 A0A0D8XVE9 A0A1B0DBF3 A0A1W0X6F6 A0A368H7B7 A0A3B0JKR2 T1JKD0 A0A261BUR5 A0A212F7F8 A0A0B7C344 A0A2G9U9M6 A0A1W4UQ62 A0A1I8CKU1 A0A0N4WL87 B0W2W3 A0A0M4ET73 O01666 W2T3Y8 B4R084 B4PPK7 B4HZE2 B3P5D7 A0A0D6M0I6 A0A016VYF3 A0A016VW22 V5I9Q2 A0A2A4JBB2 V3ZRP6 A0A2W1B7N2 A0A3B0EA27 A0A286Q4X0 T1PF90 B4JYM7 A0A286Q4W0 A0A2A2JGN9 A0A286Q4W4 E3N566 A0A2J7PRF8 W8BR16 A0A0N5AE83 A0A067RHY8 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 B8RJB4 A0A0K8U3G5 J3JTS0
A0A212EKD8 A0A194PLV8 A0A194QZB7 A0A1Y1LVM0 A0A1I8C016 A0A0V1JZ94 A0A0V0YCW8 A0A0V1NT42 A0A1Y3EIA8 A0A0V0VAI0 A0A0V0WD05 A0A0V0UDS0 A0A0V1A3F7 A0A0V1CQD4 A0A1D2N7T7 A0A267G1M7 A0A1I8HV25 E5RZD5 A0A0V1EVZ2 A0A1I8H306 A0A0V1L6C9 A0A0V1BZ30 C1BPC0 A0A210Q163 A0A0V1MFY0 A0A0U2M8V4 A0A0V1GYS8 A0A158PEG7 A0A3G1T1C7 A0A183FFX2 A0A3P7Y210 G9D519 A0A2H1WBL5 G9D526 S4NNE5 A0A077ZAI2 Q1HPX4 A0A0N4XXV6 A0A1D1UVH4 B6V839 R7UAF7 A0A085MJX5 A0A0N5E5B1 A0A0K2T9M4 A0A3M7SU40 C1BTJ1 A0A1V9XES2 A0A0D8XVE9 A0A1B0DBF3 A0A1W0X6F6 A0A368H7B7 A0A3B0JKR2 T1JKD0 A0A261BUR5 A0A212F7F8 A0A0B7C344 A0A2G9U9M6 A0A1W4UQ62 A0A1I8CKU1 A0A0N4WL87 B0W2W3 A0A0M4ET73 O01666 W2T3Y8 B4R084 B4PPK7 B4HZE2 B3P5D7 A0A0D6M0I6 A0A016VYF3 A0A016VW22 V5I9Q2 A0A2A4JBB2 V3ZRP6 A0A2W1B7N2 A0A3B0EA27 A0A286Q4X0 T1PF90 B4JYM7 A0A286Q4W0 A0A2A2JGN9 A0A286Q4W4 E3N566 A0A2J7PRF8 W8BR16 A0A0N5AE83 A0A067RHY8 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 B8RJB4 A0A0K8U3G5 J3JTS0
Pubmed
19121390
26227816
28756777
22118469
26354079
28004739
+ More
18809916 27289101 26392545 28812685 26621068 21967427 23622113 27649274 23254933 24929829 30375419 28327890 26856411 26829753 10731132 12537572 12537569 10071211 17994087 17550304 25730766 29021169 25315136 26114425 26394399 24495485 24845553 25348373 22516182 23537049
18809916 27289101 26392545 28812685 26621068 21967427 23622113 27649274 23254933 24929829 30375419 28327890 26856411 26829753 10731132 12537572 12537569 10071211 17994087 17550304 25730766 29021169 25315136 26114425 26394399 24495485 24845553 25348373 22516182 23537049
EMBL
BABH01031222
BABH01031223
JTDY01002019
KOB72310.1
ODYU01003024
SOQ41271.1
+ More
NWSH01000836 PCG73933.1 PCG73932.1 KZ150031 PZC74730.1 AGBW02014274 OWR41946.1 KQ459599 KPI94312.1 KQ460953 KPJ10320.1 GEZM01050230 GEZM01050229 JAV75666.1 JYDU01000024 JYDT01000013 JYDV01000028 KRX98119.1 KRY91455.1 KRZ40305.1 JYDS01000042 KRX98120.1 KRZ29729.1 JYDM01000105 KRZ87152.1 LVZM01014488 OUC43576.1 JYDN01000058 KRX60504.1 JYDK01000172 KRX73568.1 JYDJ01000016 KRX49494.1 JYDQ01000037 KRY19314.1 JYDI01000127 KRY51382.1 LJIJ01000160 ODN01324.1 NIVC01000639 PAA79354.1 JYDR01000005 KRY78040.1 NIVC01001626 NIVC01000386 PAA65716.1 PAA84168.1 JYDW01000122 KRZ55107.1 JYDH01000004 KRY42319.1 BT076449 ACO10873.1 NEDP02005265 OWF42490.1 JYDO01000107 KRZ70811.1 KT754301 ALS04135.1 JYDP01000196 KRZ03321.1 UYYA01000338 VDM53554.1 MG846921 AXY94773.1 UZAH01025477 VDO64704.1 JF777422 AEU11767.1 ODYU01007571 SOQ50453.1 JF777429 AEU11774.1 GAIX01012289 JAA80271.1 HG805971 CDW55680.1 BABH01018421 DQ443278 ABF51367.1 UYSL01019934 VDL71451.1 BDGG01000002 GAU93686.1 FJ267591 ACI49708.1 AMQN01001507 KB303456 ELU03111.1 KL363188 KL367571 KFD57521.1 KFD63556.1 HACA01005134 CDW22495.1 REGN01000772 RNA39219.1 BT077920 BT120664 ACO12344.1 ADD24304.1 MNPL01013186 OQR71903.1 KN716261 KJH48618.1 AJVK01029934 MTYJ01000013 OQV23146.1 JOJR01000016 RCN51187.1 OUUW01000007 SPP82914.1 JH431830 NIPN01000072 OZG14062.1 AGBW02009877 OWR49666.1 HACG01051984 CEK98855.1 KZ347909 PIO66966.1 UZAF01017700 VDO44148.1 DS231829 EDS30322.1 CP012526 ALC48060.1 AE014297 AY113454 Y12701 KI660262 ETN75946.1 CM000364 EDX14890.1 CM000160 EDW98257.1 CH480819 EDW53399.1 CH954182 EDV53187.1 KE124828 EPB77775.1 JARK01001339 EYC31808.1 EYC31809.1 GALX01002540 JAB65926.1 NWSH01002112 PCG69139.1 KB203301 ESO85225.1 KZ150244 PZC71889.1 RBVL01000238 RKO10274.1 KU728793 AOR07090.1 KA647354 AFP61983.1 CH916377 EDV90789.1 KU728788 AOR07085.1 LIAE01010453 PAV60739.1 KU728791 AOR07088.1 DS268530 DS268575 NMWX01000006 LFJK02000008 EFO87043.1 EFO91108.1 OZF97601.1 POM45268.1 NEVH01022362 PNF18917.1 GAMC01010884 JAB95671.1 KK852622 KDR19994.1 GDHF01012143 JAI40171.1 GAKP01006469 JAC52483.1 GBYB01006053 JAG75820.1 EZ000539 ACJ64413.1 GDHF01031075 JAI21239.1 APGK01025935 BT126629 KB740605 KB632095 AEE61592.1 ENN80061.1 ERL88735.1
NWSH01000836 PCG73933.1 PCG73932.1 KZ150031 PZC74730.1 AGBW02014274 OWR41946.1 KQ459599 KPI94312.1 KQ460953 KPJ10320.1 GEZM01050230 GEZM01050229 JAV75666.1 JYDU01000024 JYDT01000013 JYDV01000028 KRX98119.1 KRY91455.1 KRZ40305.1 JYDS01000042 KRX98120.1 KRZ29729.1 JYDM01000105 KRZ87152.1 LVZM01014488 OUC43576.1 JYDN01000058 KRX60504.1 JYDK01000172 KRX73568.1 JYDJ01000016 KRX49494.1 JYDQ01000037 KRY19314.1 JYDI01000127 KRY51382.1 LJIJ01000160 ODN01324.1 NIVC01000639 PAA79354.1 JYDR01000005 KRY78040.1 NIVC01001626 NIVC01000386 PAA65716.1 PAA84168.1 JYDW01000122 KRZ55107.1 JYDH01000004 KRY42319.1 BT076449 ACO10873.1 NEDP02005265 OWF42490.1 JYDO01000107 KRZ70811.1 KT754301 ALS04135.1 JYDP01000196 KRZ03321.1 UYYA01000338 VDM53554.1 MG846921 AXY94773.1 UZAH01025477 VDO64704.1 JF777422 AEU11767.1 ODYU01007571 SOQ50453.1 JF777429 AEU11774.1 GAIX01012289 JAA80271.1 HG805971 CDW55680.1 BABH01018421 DQ443278 ABF51367.1 UYSL01019934 VDL71451.1 BDGG01000002 GAU93686.1 FJ267591 ACI49708.1 AMQN01001507 KB303456 ELU03111.1 KL363188 KL367571 KFD57521.1 KFD63556.1 HACA01005134 CDW22495.1 REGN01000772 RNA39219.1 BT077920 BT120664 ACO12344.1 ADD24304.1 MNPL01013186 OQR71903.1 KN716261 KJH48618.1 AJVK01029934 MTYJ01000013 OQV23146.1 JOJR01000016 RCN51187.1 OUUW01000007 SPP82914.1 JH431830 NIPN01000072 OZG14062.1 AGBW02009877 OWR49666.1 HACG01051984 CEK98855.1 KZ347909 PIO66966.1 UZAF01017700 VDO44148.1 DS231829 EDS30322.1 CP012526 ALC48060.1 AE014297 AY113454 Y12701 KI660262 ETN75946.1 CM000364 EDX14890.1 CM000160 EDW98257.1 CH480819 EDW53399.1 CH954182 EDV53187.1 KE124828 EPB77775.1 JARK01001339 EYC31808.1 EYC31809.1 GALX01002540 JAB65926.1 NWSH01002112 PCG69139.1 KB203301 ESO85225.1 KZ150244 PZC71889.1 RBVL01000238 RKO10274.1 KU728793 AOR07090.1 KA647354 AFP61983.1 CH916377 EDV90789.1 KU728788 AOR07085.1 LIAE01010453 PAV60739.1 KU728791 AOR07088.1 DS268530 DS268575 NMWX01000006 LFJK02000008 EFO87043.1 EFO91108.1 OZF97601.1 POM45268.1 NEVH01022362 PNF18917.1 GAMC01010884 JAB95671.1 KK852622 KDR19994.1 GDHF01012143 JAI40171.1 GAKP01006469 JAC52483.1 GBYB01006053 JAG75820.1 EZ000539 ACJ64413.1 GDHF01031075 JAI21239.1 APGK01025935 BT126629 KB740605 KB632095 AEE61592.1 ENN80061.1 ERL88735.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000095281 UP000054815 UP000054826 UP000054995 UP000054805 UP000054924 UP000243006 UP000054681 UP000054673 UP000055048 UP000054783 UP000054653 UP000094527 UP000215902 UP000095280 UP000054632 UP000054721 UP000054776 UP000242188 UP000054843 UP000055024 UP000050601 UP000267027 UP000050761 UP000030665 UP000038043 UP000271162 UP000186922 UP000014760 UP000046395 UP000276133 UP000192247 UP000053766 UP000092462 UP000252519 UP000268350 UP000216463 UP000192221 UP000095286 UP000038042 UP000268014 UP000002320 UP000092553 UP000000803 UP000000304 UP000002282 UP000001292 UP000008711 UP000024635 UP000030746 UP000276569 UP000095301 UP000001070 UP000218231 UP000008281 UP000216624 UP000237256 UP000235965 UP000046393 UP000027135 UP000019118 UP000030742
UP000095281 UP000054815 UP000054826 UP000054995 UP000054805 UP000054924 UP000243006 UP000054681 UP000054673 UP000055048 UP000054783 UP000054653 UP000094527 UP000215902 UP000095280 UP000054632 UP000054721 UP000054776 UP000242188 UP000054843 UP000055024 UP000050601 UP000267027 UP000050761 UP000030665 UP000038043 UP000271162 UP000186922 UP000014760 UP000046395 UP000276133 UP000192247 UP000053766 UP000092462 UP000252519 UP000268350 UP000216463 UP000192221 UP000095286 UP000038042 UP000268014 UP000002320 UP000092553 UP000000803 UP000000304 UP000002282 UP000001292 UP000008711 UP000024635 UP000030746 UP000276569 UP000095301 UP000001070 UP000218231 UP000008281 UP000216624 UP000237256 UP000235965 UP000046393 UP000027135 UP000019118 UP000030742
Pfam
PF00231 ATP-synt
Interpro
SUPFAM
SSF52943
SSF52943
ProteinModelPortal
H9J302
A0A0L7LA84
A0A2H1VKF2
A0A2A4JR52
A0A2A4JR83
A0A2W1BPS2
+ More
A0A212EKD8 A0A194PLV8 A0A194QZB7 A0A1Y1LVM0 A0A1I8C016 A0A0V1JZ94 A0A0V0YCW8 A0A0V1NT42 A0A1Y3EIA8 A0A0V0VAI0 A0A0V0WD05 A0A0V0UDS0 A0A0V1A3F7 A0A0V1CQD4 A0A1D2N7T7 A0A267G1M7 A0A1I8HV25 E5RZD5 A0A0V1EVZ2 A0A1I8H306 A0A0V1L6C9 A0A0V1BZ30 C1BPC0 A0A210Q163 A0A0V1MFY0 A0A0U2M8V4 A0A0V1GYS8 A0A158PEG7 A0A3G1T1C7 A0A183FFX2 A0A3P7Y210 G9D519 A0A2H1WBL5 G9D526 S4NNE5 A0A077ZAI2 Q1HPX4 A0A0N4XXV6 A0A1D1UVH4 B6V839 R7UAF7 A0A085MJX5 A0A0N5E5B1 A0A0K2T9M4 A0A3M7SU40 C1BTJ1 A0A1V9XES2 A0A0D8XVE9 A0A1B0DBF3 A0A1W0X6F6 A0A368H7B7 A0A3B0JKR2 T1JKD0 A0A261BUR5 A0A212F7F8 A0A0B7C344 A0A2G9U9M6 A0A1W4UQ62 A0A1I8CKU1 A0A0N4WL87 B0W2W3 A0A0M4ET73 O01666 W2T3Y8 B4R084 B4PPK7 B4HZE2 B3P5D7 A0A0D6M0I6 A0A016VYF3 A0A016VW22 V5I9Q2 A0A2A4JBB2 V3ZRP6 A0A2W1B7N2 A0A3B0EA27 A0A286Q4X0 T1PF90 B4JYM7 A0A286Q4W0 A0A2A2JGN9 A0A286Q4W4 E3N566 A0A2J7PRF8 W8BR16 A0A0N5AE83 A0A067RHY8 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 B8RJB4 A0A0K8U3G5 J3JTS0
A0A212EKD8 A0A194PLV8 A0A194QZB7 A0A1Y1LVM0 A0A1I8C016 A0A0V1JZ94 A0A0V0YCW8 A0A0V1NT42 A0A1Y3EIA8 A0A0V0VAI0 A0A0V0WD05 A0A0V0UDS0 A0A0V1A3F7 A0A0V1CQD4 A0A1D2N7T7 A0A267G1M7 A0A1I8HV25 E5RZD5 A0A0V1EVZ2 A0A1I8H306 A0A0V1L6C9 A0A0V1BZ30 C1BPC0 A0A210Q163 A0A0V1MFY0 A0A0U2M8V4 A0A0V1GYS8 A0A158PEG7 A0A3G1T1C7 A0A183FFX2 A0A3P7Y210 G9D519 A0A2H1WBL5 G9D526 S4NNE5 A0A077ZAI2 Q1HPX4 A0A0N4XXV6 A0A1D1UVH4 B6V839 R7UAF7 A0A085MJX5 A0A0N5E5B1 A0A0K2T9M4 A0A3M7SU40 C1BTJ1 A0A1V9XES2 A0A0D8XVE9 A0A1B0DBF3 A0A1W0X6F6 A0A368H7B7 A0A3B0JKR2 T1JKD0 A0A261BUR5 A0A212F7F8 A0A0B7C344 A0A2G9U9M6 A0A1W4UQ62 A0A1I8CKU1 A0A0N4WL87 B0W2W3 A0A0M4ET73 O01666 W2T3Y8 B4R084 B4PPK7 B4HZE2 B3P5D7 A0A0D6M0I6 A0A016VYF3 A0A016VW22 V5I9Q2 A0A2A4JBB2 V3ZRP6 A0A2W1B7N2 A0A3B0EA27 A0A286Q4X0 T1PF90 B4JYM7 A0A286Q4W0 A0A2A2JGN9 A0A286Q4W4 E3N566 A0A2J7PRF8 W8BR16 A0A0N5AE83 A0A067RHY8 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 B8RJB4 A0A0K8U3G5 J3JTS0
PDB
2W6J
E-value=1.60021e-12,
Score=169
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion
Mitochondrion inner membrane
Mitochondrion inner membrane
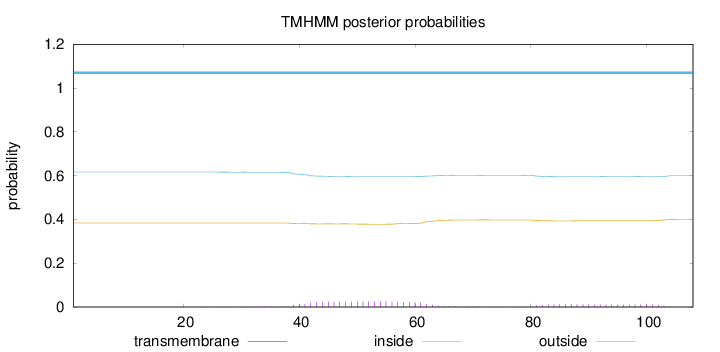
Length:
108
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.83268
Exp number, first 60 AAs:
0.50699
Total prob of N-in:
0.61680
inside
1 - 108
Population Genetic Test Statistics
Pi
7.950628
Theta
9.030718
Tajima's D
-0.341113
CLR
0
CSRT
0.291385430728464
Interpretation
Uncertain
