Gene
KWMTBOMO00647
Pre Gene Modal
BGIBMGA003889
Annotation
PREDICTED:_ATP_synthase_subunit_gamma?_mitochondrial-like_[Amyelois_transitella]
Full name
ATP synthase subunit gamma
Location in the cell
Cytoplasmic Reliability : 1.131 Nuclear Reliability : 1.209
Sequence
CDS
ATGACAGCCTTCGATGACGTGGATGACGACCAGCTGGAGTGCTACGCGGAGTGGACCCTCGCCGCGCTCCTCTACTACGCGTTGAAGCAAGGCGCGGCTTCAGAACAGTCTGCCCGGATGGCTGCCATGGACAACGCTACGAAGAACGCATCCGAAATGATCAGGAAACTGACGCTGCTCTTCAACAGAACGAGACAAGCAGTCATCACCAGGGAGCTTATTGAAATCATTTCAGGAGCGGCGGCTTTAAAGTGA
Protein
MTAFDDVDDDQLECYAEWTLAALLYYALKQGAASEQSARMAAMDNATKNASEMIRKLTLLFNRTRQAVITRELIEIISGAAALK
Summary
Subunit
F-type ATPases have 2 components, CF(1) - the catalytic core - and CF(0) - the membrane proton channel. CF(1) has five subunits: alpha(3), beta(3), gamma(1), delta(1), epsilon(1). CF(0) has three main subunits: a, b and c.
Similarity
Belongs to the ATPase gamma chain family.
Uniprot
H9J302
A0A2W1BPS2
A0A2A4JR52
A0A2A4JR83
A0A194PLV8
A0A194QZB7
+ More
A0A2H1VKF2 A0A212EKD8 A0A0L7LA84 A0A091EGE1 U3IK89 F6SJM8 A0A1X7VD82 S9W3T2 Q1HPX4 K7P5E1 A0A0P6CI98 A0A2G9S1E0 A0A1D1ZG56 A0A2G9S1B1 U6DD18 A0A0P4VV79 R4FNB9 K7P534 T1HN70 L5LBI1 A0A1S3EKA9 A0A3L7I0Q9 A0A2I0T2W2 A0A3M6U7F3 F4WG50 F7F7L6 A0A1L1RPA5 L9KQG5 A0A023F8R4 A0A0C9QPL6 L5L0J2 G1NXI0 C1C3J8 A0A069DRH5 D6WPW7 G3TDA5 A0A341AG22 A0A2U4B6F4 S7NQV4 G3GR61 A0A2U3YCA8 K7P595 A0A0L7LUJ4 A0A2K6MWG2 A0A250YEF4 I4DJ96 S4NNE5 Q4R7P5 A0A2Y9HL94 R0LGI8 A0A2S2R2P5 D2I3D1 A0A1S3AFZ6 A0A2Y9HSU1 A0A2Y9R6L4 G1SM77 A0A2Y9H1B6 A0A0V0G371 A0A2U3ZDN1 B4DL14 A0A1L1RQ30 A0A212F7F8 A0A2U3V799 A0A340XXI4 A0A224XPP2 A0A1D5PXH6 A0A0K0DQ69 A0A2Y9NLH3 H0VH31 K9IZN0 A0A2Y9KRW5 A0A2U3V6X1 A0A340Y055 A0A2R7WX91 K9KF86 A0A341ABI7 A0A286XFY1 A0A2Y9NY33 A0A0N4TC37 A0A2K6D1F5 A0A2K6D1F4 A0A341ABQ1 A0A3Q0DE26 A0A2K6N6Z9 A0A3M0KGV2 A0A2K6FE13 A0A3Q0DL77 A0A2Y9KYQ2 A8Y2E9 A0A0A9Z7I7 E3N566 A0A261BUR5 A0A2K6N702 A0A026WWE4
A0A2H1VKF2 A0A212EKD8 A0A0L7LA84 A0A091EGE1 U3IK89 F6SJM8 A0A1X7VD82 S9W3T2 Q1HPX4 K7P5E1 A0A0P6CI98 A0A2G9S1E0 A0A1D1ZG56 A0A2G9S1B1 U6DD18 A0A0P4VV79 R4FNB9 K7P534 T1HN70 L5LBI1 A0A1S3EKA9 A0A3L7I0Q9 A0A2I0T2W2 A0A3M6U7F3 F4WG50 F7F7L6 A0A1L1RPA5 L9KQG5 A0A023F8R4 A0A0C9QPL6 L5L0J2 G1NXI0 C1C3J8 A0A069DRH5 D6WPW7 G3TDA5 A0A341AG22 A0A2U4B6F4 S7NQV4 G3GR61 A0A2U3YCA8 K7P595 A0A0L7LUJ4 A0A2K6MWG2 A0A250YEF4 I4DJ96 S4NNE5 Q4R7P5 A0A2Y9HL94 R0LGI8 A0A2S2R2P5 D2I3D1 A0A1S3AFZ6 A0A2Y9HSU1 A0A2Y9R6L4 G1SM77 A0A2Y9H1B6 A0A0V0G371 A0A2U3ZDN1 B4DL14 A0A1L1RQ30 A0A212F7F8 A0A2U3V799 A0A340XXI4 A0A224XPP2 A0A1D5PXH6 A0A0K0DQ69 A0A2Y9NLH3 H0VH31 K9IZN0 A0A2Y9KRW5 A0A2U3V6X1 A0A340Y055 A0A2R7WX91 K9KF86 A0A341ABI7 A0A286XFY1 A0A2Y9NY33 A0A0N4TC37 A0A2K6D1F5 A0A2K6D1F4 A0A341ABQ1 A0A3Q0DE26 A0A2K6N6Z9 A0A3M0KGV2 A0A2K6FE13 A0A3Q0DL77 A0A2Y9KYQ2 A8Y2E9 A0A0A9Z7I7 E3N566 A0A261BUR5 A0A2K6N702 A0A026WWE4
Pubmed
19121390
28756777
26354079
22118469
26227816
23749191
+ More
19892987 23149746 22438331 27129103 29704459 30382153 21719571 18464734 23385571 25474469 23258410 21993624 26334808 18362917 19820115 21804562 28087693 22651552 23622113 15944441 20010809 25362486 14624247 25401762 26823975 26114425 26394399 24508170 30249741
19892987 23149746 22438331 27129103 29704459 30382153 21719571 18464734 23385571 25474469 23258410 21993624 26334808 18362917 19820115 21804562 28087693 22651552 23622113 15944441 20010809 25362486 14624247 25401762 26823975 26114425 26394399 24508170 30249741
EMBL
BABH01031222
BABH01031223
KZ150031
PZC74730.1
NWSH01000836
PCG73933.1
+ More
PCG73932.1 KQ459599 KPI94312.1 KQ460953 KPJ10320.1 ODYU01003024 SOQ41271.1 AGBW02014274 OWR41946.1 JTDY01002019 KOB72310.1 KN121943 KFO34506.1 ADON01089360 ADON01089361 ADON01089362 KB020093 EPY72642.1 BABH01018421 DQ443278 ABF51367.1 JU062556 AFC92867.1 GDIP01001512 JAN02204.1 KV927954 PIO33875.1 GDJX01002097 JAT65839.1 PIO33874.1 HAAF01009026 CCP80850.1 GDKW01000647 JAI55948.1 GAHY01001359 JAA76151.1 JU062554 AFC92865.1 ACPB03017427 KB113569 ELK23537.1 RAZU01000120 RLQ71713.1 KZ522286 PKU28136.1 RCHS01002096 RMX49557.1 GL888128 EGI66817.1 KB320704 ELW64996.1 GBBI01000906 JAC17806.1 GBYB01005564 JAG75331.1 KB030418 ELK16910.1 AAPE02016991 AAPE02016992 BT081427 ACO51558.1 GBGD01002379 JAC86510.1 KQ971354 EFA07448.1 KE164710 EPQ19646.1 JH000001 EGV95073.1 JU062555 AFC92866.1 JTDY01000057 KOB79120.1 GFFW01002854 JAV41934.1 AK401364 KQ459583 BAM17986.1 KPI98617.1 GAIX01012289 JAA80271.1 AB168770 BAE00877.1 KB743338 EOA99357.1 GGMS01014847 MBY84050.1 GL194296 EFB18250.1 AAGW02026845 AAGW02026846 GECL01003595 JAP02529.1 AK296803 BAG59376.1 AGBW02009877 OWR49666.1 GFTR01004628 JAW11798.1 AAKN02021916 AAKN02021917 GABZ01007211 JAA46314.1 KK855861 PTY24152.1 JL626894 AEQ00022.1 UZAD01004376 UZAD01006801 VDN86924.1 VDN88006.1 QRBI01000112 RMC10484.1 HE600977 CAP39071.1 GBHO01004281 GDHC01013990 JAG39323.1 JAQ04639.1 DS268530 DS268575 NMWX01000006 LFJK02000008 EFO87043.1 EFO91108.1 OZF97601.1 POM45268.1 NIPN01000072 OZG14062.1 KK107085 QOIP01000008 EZA60056.1 RLU19527.1
PCG73932.1 KQ459599 KPI94312.1 KQ460953 KPJ10320.1 ODYU01003024 SOQ41271.1 AGBW02014274 OWR41946.1 JTDY01002019 KOB72310.1 KN121943 KFO34506.1 ADON01089360 ADON01089361 ADON01089362 KB020093 EPY72642.1 BABH01018421 DQ443278 ABF51367.1 JU062556 AFC92867.1 GDIP01001512 JAN02204.1 KV927954 PIO33875.1 GDJX01002097 JAT65839.1 PIO33874.1 HAAF01009026 CCP80850.1 GDKW01000647 JAI55948.1 GAHY01001359 JAA76151.1 JU062554 AFC92865.1 ACPB03017427 KB113569 ELK23537.1 RAZU01000120 RLQ71713.1 KZ522286 PKU28136.1 RCHS01002096 RMX49557.1 GL888128 EGI66817.1 KB320704 ELW64996.1 GBBI01000906 JAC17806.1 GBYB01005564 JAG75331.1 KB030418 ELK16910.1 AAPE02016991 AAPE02016992 BT081427 ACO51558.1 GBGD01002379 JAC86510.1 KQ971354 EFA07448.1 KE164710 EPQ19646.1 JH000001 EGV95073.1 JU062555 AFC92866.1 JTDY01000057 KOB79120.1 GFFW01002854 JAV41934.1 AK401364 KQ459583 BAM17986.1 KPI98617.1 GAIX01012289 JAA80271.1 AB168770 BAE00877.1 KB743338 EOA99357.1 GGMS01014847 MBY84050.1 GL194296 EFB18250.1 AAGW02026845 AAGW02026846 GECL01003595 JAP02529.1 AK296803 BAG59376.1 AGBW02009877 OWR49666.1 GFTR01004628 JAW11798.1 AAKN02021916 AAKN02021917 GABZ01007211 JAA46314.1 KK855861 PTY24152.1 JL626894 AEQ00022.1 UZAD01004376 UZAD01006801 VDN86924.1 VDN88006.1 QRBI01000112 RMC10484.1 HE600977 CAP39071.1 GBHO01004281 GDHC01013990 JAG39323.1 JAQ04639.1 DS268530 DS268575 NMWX01000006 LFJK02000008 EFO87043.1 EFO91108.1 OZF97601.1 POM45268.1 NIPN01000072 OZG14062.1 KK107085 QOIP01000008 EZA60056.1 RLU19527.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000028990 UP000016666 UP000002281 UP000007879 UP000015103 UP000081671 UP000273346 UP000275408 UP000007755 UP000002279 UP000011518 UP000010552 UP000001074 UP000007266 UP000007646 UP000252040 UP000245320 UP000001075 UP000245341 UP000233180 UP000248481 UP000079721 UP000248480 UP000001811 UP000245340 UP000265300 UP000035642 UP000248483 UP000005447 UP000248482 UP000038020 UP000278627 UP000233120 UP000189704 UP000233200 UP000269221 UP000233160 UP000008549 UP000008281 UP000216624 UP000237256 UP000216463 UP000053097 UP000279307
UP000028990 UP000016666 UP000002281 UP000007879 UP000015103 UP000081671 UP000273346 UP000275408 UP000007755 UP000002279 UP000011518 UP000010552 UP000001074 UP000007266 UP000007646 UP000252040 UP000245320 UP000001075 UP000245341 UP000233180 UP000248481 UP000079721 UP000248480 UP000001811 UP000245340 UP000265300 UP000035642 UP000248483 UP000005447 UP000248482 UP000038020 UP000278627 UP000233120 UP000189704 UP000233200 UP000269221 UP000233160 UP000008549 UP000008281 UP000216624 UP000237256 UP000216463 UP000053097 UP000279307
Interpro
Gene 3D
ProteinModelPortal
H9J302
A0A2W1BPS2
A0A2A4JR52
A0A2A4JR83
A0A194PLV8
A0A194QZB7
+ More
A0A2H1VKF2 A0A212EKD8 A0A0L7LA84 A0A091EGE1 U3IK89 F6SJM8 A0A1X7VD82 S9W3T2 Q1HPX4 K7P5E1 A0A0P6CI98 A0A2G9S1E0 A0A1D1ZG56 A0A2G9S1B1 U6DD18 A0A0P4VV79 R4FNB9 K7P534 T1HN70 L5LBI1 A0A1S3EKA9 A0A3L7I0Q9 A0A2I0T2W2 A0A3M6U7F3 F4WG50 F7F7L6 A0A1L1RPA5 L9KQG5 A0A023F8R4 A0A0C9QPL6 L5L0J2 G1NXI0 C1C3J8 A0A069DRH5 D6WPW7 G3TDA5 A0A341AG22 A0A2U4B6F4 S7NQV4 G3GR61 A0A2U3YCA8 K7P595 A0A0L7LUJ4 A0A2K6MWG2 A0A250YEF4 I4DJ96 S4NNE5 Q4R7P5 A0A2Y9HL94 R0LGI8 A0A2S2R2P5 D2I3D1 A0A1S3AFZ6 A0A2Y9HSU1 A0A2Y9R6L4 G1SM77 A0A2Y9H1B6 A0A0V0G371 A0A2U3ZDN1 B4DL14 A0A1L1RQ30 A0A212F7F8 A0A2U3V799 A0A340XXI4 A0A224XPP2 A0A1D5PXH6 A0A0K0DQ69 A0A2Y9NLH3 H0VH31 K9IZN0 A0A2Y9KRW5 A0A2U3V6X1 A0A340Y055 A0A2R7WX91 K9KF86 A0A341ABI7 A0A286XFY1 A0A2Y9NY33 A0A0N4TC37 A0A2K6D1F5 A0A2K6D1F4 A0A341ABQ1 A0A3Q0DE26 A0A2K6N6Z9 A0A3M0KGV2 A0A2K6FE13 A0A3Q0DL77 A0A2Y9KYQ2 A8Y2E9 A0A0A9Z7I7 E3N566 A0A261BUR5 A0A2K6N702 A0A026WWE4
A0A2H1VKF2 A0A212EKD8 A0A0L7LA84 A0A091EGE1 U3IK89 F6SJM8 A0A1X7VD82 S9W3T2 Q1HPX4 K7P5E1 A0A0P6CI98 A0A2G9S1E0 A0A1D1ZG56 A0A2G9S1B1 U6DD18 A0A0P4VV79 R4FNB9 K7P534 T1HN70 L5LBI1 A0A1S3EKA9 A0A3L7I0Q9 A0A2I0T2W2 A0A3M6U7F3 F4WG50 F7F7L6 A0A1L1RPA5 L9KQG5 A0A023F8R4 A0A0C9QPL6 L5L0J2 G1NXI0 C1C3J8 A0A069DRH5 D6WPW7 G3TDA5 A0A341AG22 A0A2U4B6F4 S7NQV4 G3GR61 A0A2U3YCA8 K7P595 A0A0L7LUJ4 A0A2K6MWG2 A0A250YEF4 I4DJ96 S4NNE5 Q4R7P5 A0A2Y9HL94 R0LGI8 A0A2S2R2P5 D2I3D1 A0A1S3AFZ6 A0A2Y9HSU1 A0A2Y9R6L4 G1SM77 A0A2Y9H1B6 A0A0V0G371 A0A2U3ZDN1 B4DL14 A0A1L1RQ30 A0A212F7F8 A0A2U3V799 A0A340XXI4 A0A224XPP2 A0A1D5PXH6 A0A0K0DQ69 A0A2Y9NLH3 H0VH31 K9IZN0 A0A2Y9KRW5 A0A2U3V6X1 A0A340Y055 A0A2R7WX91 K9KF86 A0A341ABI7 A0A286XFY1 A0A2Y9NY33 A0A0N4TC37 A0A2K6D1F5 A0A2K6D1F4 A0A341ABQ1 A0A3Q0DE26 A0A2K6N6Z9 A0A3M0KGV2 A0A2K6FE13 A0A3Q0DL77 A0A2Y9KYQ2 A8Y2E9 A0A0A9Z7I7 E3N566 A0A261BUR5 A0A2K6N702 A0A026WWE4
PDB
2W6J
E-value=4.45639e-25,
Score=277
Ontologies
GO
PANTHER
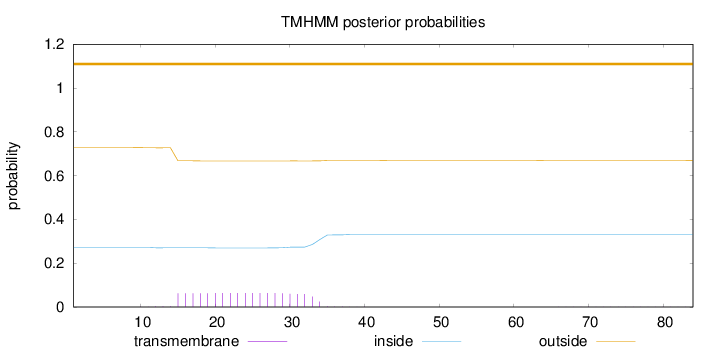
Topology
Length:
84
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.21793
Exp number, first 60 AAs:
1.21167
Total prob of N-in:
0.27193
outside
1 - 84
Population Genetic Test Statistics
Pi
16.149616
Theta
27.403213
Tajima's D
-1.261734
CLR
0.451045
CSRT
0.0897455127243638
Interpretation
Uncertain
