Gene
KWMTBOMO00644
Pre Gene Modal
BGIBMGA003864
Annotation
os_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.974
Sequence
CDS
ATGTCGAAGAACGGTTTAGAGGAACCAAGCGGCAGTCAGCCGACGCCGAAGGAGACTACGCCGCTCATTACAAAGGGTGGCGAAAATGGCAACGGTGGTGGTAAGAACGGCGGACTCAGCGTTGGGCAGACGAGTATGTTGATCGCCGGAGAGATGGCAGGCAGCGGAGTCTTGGCTCTCCCTAGAGCCCTTGTCAGGACTGGATGGATTGGTGTTCCCATTATCATTCTGATGTGCTTGGTGGCGGCCTTCAGTGGCAAGCGACTCGGTGACTGCTGGACCATTCTGGAGGAACGCAATCCTCAACTGAGGAGCAGGAAGAGGAACCCATACGCTATTATTGCTGATCAAACTCTCGGCAAGACCTGGAGCGTGGTCGTGTCCATGGCGATCATTGTAACCCTCTTTGGAGCTTCGGTCGTGTACCTGCTGATGGCAGCTCAGATCATCGAACAACTCCTACTCACTCTGATCCCAACATTGACCATTTGCACTTGGTACCTGATCGTGGTCGGAGCCATGACTCCTCTTATCTTCTTTAACAGCCCTAAGGATCTGACCTTCACTGGTGTAATCGCCTTCGGATCCACGGTGATTGCCTGCATCTTGTACTTTATTGAGATGATGAACGAAGTCCGTCCTTTCGTGTTTCGTTGGGGTGTCCACGGCTTTACGGATTTCTTCCTTGCTTTCGGCACCATCATGTTTGCGTTTGGGGGTGCCTCAACCTTTCCAACCATCCAGAACGACATGACCGACAAATCTCAGTTTGGGAAAAGCATTCAGTACAGTTTTGGGGCCATCCTGTTGCTGTACCTACCCATCGCGATCGGAGGCTATGCTGTGTACGGCGAGTCAGTTGGGTCTAACGTGGCCTTATCTCTGTCTGCTACCCCGTTGACCCTGGTCGGAAACATCTTTATGGCCATTCACCTCGTCTTCGCTTTTATCATCTTGATTAACCCGGTCTGCCAGGAAATGGAAGAGATTTACAACATTGAGAGAGATTCCGTGGGCTGGCGTGTCCTGATCCGCCTCTCCATCATGGGCGCCATTTTGTTCATCGGCGAGAGTATTCCTCGTTTCTACACGATCCTCGCGCTGGTCGGAGGCACAACAGTGGCTCTGCTCACTTACATCCTGCCTTCGTTCTGCTACCTGAGCCTCATTAACCAGACGCCTAGAGAGGGGCAGACTCCAATAGAAACTCCTGGCTGGGTGAAATTGCTTTGCTACGAGGTTATTGCCCTCGGGGTCCTTGGGGCCGTGGCCGCAACCTACAGCGGACTAAGCGCCGTGTTCAGCAGCGCCGTCACCACGCCCTGCTACCTACGATAG
Protein
MSKNGLEEPSGSQPTPKETTPLITKGGENGNGGGKNGGLSVGQTSMLIAGEMAGSGVLALPRALVRTGWIGVPIIILMCLVAAFSGKRLGDCWTILEERNPQLRSRKRNPYAIIADQTLGKTWSVVVSMAIIVTLFGASVVYLLMAAQIIEQLLLTLIPTLTICTWYLIVVGAMTPLIFFNSPKDLTFTGVIAFGSTVIACILYFIEMMNEVRPFVFRWGVHGFTDFFLAFGTIMFAFGGASTFPTIQNDMTDKSQFGKSIQYSFGAILLLYLPIAIGGYAVYGESVGSNVALSLSATPLTLVGNIFMAIHLVFAFIILINPVCQEMEEIYNIERDSVGWRVLIRLSIMGAILFIGESIPRFYTILALVGGTTVALLTYILPSFCYLSLINQTPREGQTPIETPGWVKLLCYEVIALGVLGAVAATYSGLSAVFSSAVTTPCYLR
Summary
Uniprot
G1UH29
G1UH32
H9J2X7
I4DJH9
I4DM23
A0A212EKD5
+ More
A0A2A4JQ93 A0A2W1BI57 G1UH31 A0A194QXQ2 A0A067QJD4 A0A1Y1NHJ2 A0A2J7QU03 A0A2J7QU19 A0A0A9ZCL0 A0A2J7QU12 A0A023F831 A0A1B6HTY9 R4G3L0 T1HU60 A0A1B6GMY9 R4WS75 A0A1B6J618 E0V9U9 A0A1B6MPJ4 A0A1B6EVQ8 C3YUS8 C3YUS7 A0A087T2R7 A0A2C9K492 A0A1S3IZ16 A0A1S3IYH0 A0A1S3KEE1 A0A1S3KF48 T1JJG5 A0A2G8KUZ9 A0A1W3JDY6 A0A2C9K468 A0A1B6DU47 A0A2C9K488 A0A2C9K491 A0A2J7QU04 A0A146LB68 A0A2T7P419 F7A4D7 K1RT88 A0A1W0WSB9 H2Z6U2 B3RRJ6 A0A369RZ19 A0A210QID1 A0A2P2HZS8 A0A0L8IE90 H2Z6U1 A0A2T7P403 A0A2L2Y9K5 A0A2R5LEF6 L7MIU8 L7MGY3 L7MH99 A0A293LGF8 T1KQS4 A0A131XHC8 A0A131YCM8 A0A131YWW9 L7MGQ4 A0A087TTD7 A0A2P2HY10 A0A183UI32 V5H7V7 A0A0P4VZI8 F6VBX7 H2Z6I0 F0J967 A0A158PNA9 A0A3P6RQ62 T1KID8 F6VBN4 T1KJ56 A0A1B6JMD9 A0A182E8I6 A0A044U870 A0A2L2YCV6 F1L368 G3MII0 A0A0N5AWU9 E4XHC6 E4YG51 A0A023GNA8 A0A023GGI8 A0A023GD66 G3MHT0 T1JTR3 A0A2S2NQH5 A0A0N4TQC4
A0A2A4JQ93 A0A2W1BI57 G1UH31 A0A194QXQ2 A0A067QJD4 A0A1Y1NHJ2 A0A2J7QU03 A0A2J7QU19 A0A0A9ZCL0 A0A2J7QU12 A0A023F831 A0A1B6HTY9 R4G3L0 T1HU60 A0A1B6GMY9 R4WS75 A0A1B6J618 E0V9U9 A0A1B6MPJ4 A0A1B6EVQ8 C3YUS8 C3YUS7 A0A087T2R7 A0A2C9K492 A0A1S3IZ16 A0A1S3IYH0 A0A1S3KEE1 A0A1S3KF48 T1JJG5 A0A2G8KUZ9 A0A1W3JDY6 A0A2C9K468 A0A1B6DU47 A0A2C9K488 A0A2C9K491 A0A2J7QU04 A0A146LB68 A0A2T7P419 F7A4D7 K1RT88 A0A1W0WSB9 H2Z6U2 B3RRJ6 A0A369RZ19 A0A210QID1 A0A2P2HZS8 A0A0L8IE90 H2Z6U1 A0A2T7P403 A0A2L2Y9K5 A0A2R5LEF6 L7MIU8 L7MGY3 L7MH99 A0A293LGF8 T1KQS4 A0A131XHC8 A0A131YCM8 A0A131YWW9 L7MGQ4 A0A087TTD7 A0A2P2HY10 A0A183UI32 V5H7V7 A0A0P4VZI8 F6VBX7 H2Z6I0 F0J967 A0A158PNA9 A0A3P6RQ62 T1KID8 F6VBN4 T1KJ56 A0A1B6JMD9 A0A182E8I6 A0A044U870 A0A2L2YCV6 F1L368 G3MII0 A0A0N5AWU9 E4XHC6 E4YG51 A0A023GNA8 A0A023GGI8 A0A023GD66 G3MHT0 T1JTR3 A0A2S2NQH5 A0A0N4TQC4
Pubmed
EMBL
AB606415
BAK64377.1
AB606418
BAK64380.1
BABH01031223
AK401447
+ More
BAM18069.1 AK402341 BAM18963.1 AGBW02014274 OWR41944.1 NWSH01000836 PCG73936.1 KZ150031 PZC74732.1 AB606417 BAK64379.1 KQ460953 KPJ10318.1 KK853424 KDR07697.1 GEZM01002091 JAV97373.1 NEVH01011192 PNF32067.1 PNF32065.1 GBHO01001430 GBHO01001428 GBRD01006036 GBRD01000363 JAG42174.1 JAG42176.1 JAG59785.1 PNF32069.1 GBBI01001292 JAC17420.1 GECU01029601 JAS78105.1 GAHY01002115 JAA75395.1 ACPB03000737 GECZ01005961 JAS63808.1 AK417542 BAN20757.1 GECU01013109 JAS94597.1 DS234996 EEB10155.1 GEBQ01002097 JAT37880.1 GECZ01027986 GECZ01011439 JAS41783.1 JAS58330.1 GG666554 EEN55979.1 EEN55978.1 KK113129 KFM59406.1 JH431912 MRZV01000356 PIK51785.1 GEDC01008104 JAS29194.1 PNF32068.1 GDHC01014287 JAQ04342.1 PZQS01000006 PVD28156.1 EAAA01002435 JH818674 EKC37816.1 MTYJ01000053 OQV18096.1 DS985243 EDV26356.1 NOWV01000117 RDD39926.1 NEDP02003514 OWF48500.1 IACF01001590 LAB67282.1 KQ415893 KOF99782.1 PVD28157.1 IAAA01011297 IAAA01011298 LAA04823.1 GGLE01003756 MBY07882.1 GACK01001831 JAA63203.1 GACK01001563 JAA63471.1 GACK01001832 JAA63202.1 GFWV01003093 MAA27823.1 CAEY01000380 GEFH01002779 JAP65802.1 GEDV01012345 JAP76212.1 GEDV01005562 JAP82995.1 GACK01001884 JAA63150.1 KK116647 KFM68376.1 IACF01000750 LAB66506.1 UYWY01019837 VDM39473.1 GANP01011304 JAB73164.1 GDRN01104869 JAI57831.1 BK007418 DAA34377.1 UYRR01031031 VDK44068.1 CAEY01000112 CAEY01000119 GECU01007361 JAT00346.1 UYRW01000963 VDK72647.1 CMVM020000172 IAAA01010513 IAAA01010514 LAA05984.1 JI170479 ADY44572.1 JO841681 AEO33298.1 FN653051 CBY10074.1 FN654511 CBY34475.1 GBBM01000099 JAC35319.1 GBBM01003310 JAC32108.1 GBBM01003312 JAC32106.1 JO841431 AEO33048.1 CAEY01000484 GGMR01006593 MBY19212.1 UZAD01013198 VDN91939.1
BAM18069.1 AK402341 BAM18963.1 AGBW02014274 OWR41944.1 NWSH01000836 PCG73936.1 KZ150031 PZC74732.1 AB606417 BAK64379.1 KQ460953 KPJ10318.1 KK853424 KDR07697.1 GEZM01002091 JAV97373.1 NEVH01011192 PNF32067.1 PNF32065.1 GBHO01001430 GBHO01001428 GBRD01006036 GBRD01000363 JAG42174.1 JAG42176.1 JAG59785.1 PNF32069.1 GBBI01001292 JAC17420.1 GECU01029601 JAS78105.1 GAHY01002115 JAA75395.1 ACPB03000737 GECZ01005961 JAS63808.1 AK417542 BAN20757.1 GECU01013109 JAS94597.1 DS234996 EEB10155.1 GEBQ01002097 JAT37880.1 GECZ01027986 GECZ01011439 JAS41783.1 JAS58330.1 GG666554 EEN55979.1 EEN55978.1 KK113129 KFM59406.1 JH431912 MRZV01000356 PIK51785.1 GEDC01008104 JAS29194.1 PNF32068.1 GDHC01014287 JAQ04342.1 PZQS01000006 PVD28156.1 EAAA01002435 JH818674 EKC37816.1 MTYJ01000053 OQV18096.1 DS985243 EDV26356.1 NOWV01000117 RDD39926.1 NEDP02003514 OWF48500.1 IACF01001590 LAB67282.1 KQ415893 KOF99782.1 PVD28157.1 IAAA01011297 IAAA01011298 LAA04823.1 GGLE01003756 MBY07882.1 GACK01001831 JAA63203.1 GACK01001563 JAA63471.1 GACK01001832 JAA63202.1 GFWV01003093 MAA27823.1 CAEY01000380 GEFH01002779 JAP65802.1 GEDV01012345 JAP76212.1 GEDV01005562 JAP82995.1 GACK01001884 JAA63150.1 KK116647 KFM68376.1 IACF01000750 LAB66506.1 UYWY01019837 VDM39473.1 GANP01011304 JAB73164.1 GDRN01104869 JAI57831.1 BK007418 DAA34377.1 UYRR01031031 VDK44068.1 CAEY01000112 CAEY01000119 GECU01007361 JAT00346.1 UYRW01000963 VDK72647.1 CMVM020000172 IAAA01010513 IAAA01010514 LAA05984.1 JI170479 ADY44572.1 JO841681 AEO33298.1 FN653051 CBY10074.1 FN654511 CBY34475.1 GBBM01000099 JAC35319.1 GBBM01003310 JAC32108.1 GBBM01003312 JAC32106.1 JO841431 AEO33048.1 CAEY01000484 GGMR01006593 MBY19212.1 UZAD01013198 VDN91939.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000027135
UP000235965
+ More
UP000015103 UP000009046 UP000001554 UP000054359 UP000076420 UP000085678 UP000230750 UP000245119 UP000008144 UP000005408 UP000007875 UP000009022 UP000253843 UP000242188 UP000053454 UP000015104 UP000050794 UP000267007 UP000036680 UP000077448 UP000271087 UP000024404 UP000046393 UP000038020 UP000278627
UP000015103 UP000009046 UP000001554 UP000054359 UP000076420 UP000085678 UP000230750 UP000245119 UP000008144 UP000005408 UP000007875 UP000009022 UP000253843 UP000242188 UP000053454 UP000015104 UP000050794 UP000267007 UP000036680 UP000077448 UP000271087 UP000024404 UP000046393 UP000038020 UP000278627
Pfam
PF01490 Aa_trans
Gene 3D
ProteinModelPortal
G1UH29
G1UH32
H9J2X7
I4DJH9
I4DM23
A0A212EKD5
+ More
A0A2A4JQ93 A0A2W1BI57 G1UH31 A0A194QXQ2 A0A067QJD4 A0A1Y1NHJ2 A0A2J7QU03 A0A2J7QU19 A0A0A9ZCL0 A0A2J7QU12 A0A023F831 A0A1B6HTY9 R4G3L0 T1HU60 A0A1B6GMY9 R4WS75 A0A1B6J618 E0V9U9 A0A1B6MPJ4 A0A1B6EVQ8 C3YUS8 C3YUS7 A0A087T2R7 A0A2C9K492 A0A1S3IZ16 A0A1S3IYH0 A0A1S3KEE1 A0A1S3KF48 T1JJG5 A0A2G8KUZ9 A0A1W3JDY6 A0A2C9K468 A0A1B6DU47 A0A2C9K488 A0A2C9K491 A0A2J7QU04 A0A146LB68 A0A2T7P419 F7A4D7 K1RT88 A0A1W0WSB9 H2Z6U2 B3RRJ6 A0A369RZ19 A0A210QID1 A0A2P2HZS8 A0A0L8IE90 H2Z6U1 A0A2T7P403 A0A2L2Y9K5 A0A2R5LEF6 L7MIU8 L7MGY3 L7MH99 A0A293LGF8 T1KQS4 A0A131XHC8 A0A131YCM8 A0A131YWW9 L7MGQ4 A0A087TTD7 A0A2P2HY10 A0A183UI32 V5H7V7 A0A0P4VZI8 F6VBX7 H2Z6I0 F0J967 A0A158PNA9 A0A3P6RQ62 T1KID8 F6VBN4 T1KJ56 A0A1B6JMD9 A0A182E8I6 A0A044U870 A0A2L2YCV6 F1L368 G3MII0 A0A0N5AWU9 E4XHC6 E4YG51 A0A023GNA8 A0A023GGI8 A0A023GD66 G3MHT0 T1JTR3 A0A2S2NQH5 A0A0N4TQC4
A0A2A4JQ93 A0A2W1BI57 G1UH31 A0A194QXQ2 A0A067QJD4 A0A1Y1NHJ2 A0A2J7QU03 A0A2J7QU19 A0A0A9ZCL0 A0A2J7QU12 A0A023F831 A0A1B6HTY9 R4G3L0 T1HU60 A0A1B6GMY9 R4WS75 A0A1B6J618 E0V9U9 A0A1B6MPJ4 A0A1B6EVQ8 C3YUS8 C3YUS7 A0A087T2R7 A0A2C9K492 A0A1S3IZ16 A0A1S3IYH0 A0A1S3KEE1 A0A1S3KF48 T1JJG5 A0A2G8KUZ9 A0A1W3JDY6 A0A2C9K468 A0A1B6DU47 A0A2C9K488 A0A2C9K491 A0A2J7QU04 A0A146LB68 A0A2T7P419 F7A4D7 K1RT88 A0A1W0WSB9 H2Z6U2 B3RRJ6 A0A369RZ19 A0A210QID1 A0A2P2HZS8 A0A0L8IE90 H2Z6U1 A0A2T7P403 A0A2L2Y9K5 A0A2R5LEF6 L7MIU8 L7MGY3 L7MH99 A0A293LGF8 T1KQS4 A0A131XHC8 A0A131YCM8 A0A131YWW9 L7MGQ4 A0A087TTD7 A0A2P2HY10 A0A183UI32 V5H7V7 A0A0P4VZI8 F6VBX7 H2Z6I0 F0J967 A0A158PNA9 A0A3P6RQ62 T1KID8 F6VBN4 T1KJ56 A0A1B6JMD9 A0A182E8I6 A0A044U870 A0A2L2YCV6 F1L368 G3MII0 A0A0N5AWU9 E4XHC6 E4YG51 A0A023GNA8 A0A023GGI8 A0A023GD66 G3MHT0 T1JTR3 A0A2S2NQH5 A0A0N4TQC4
Ontologies
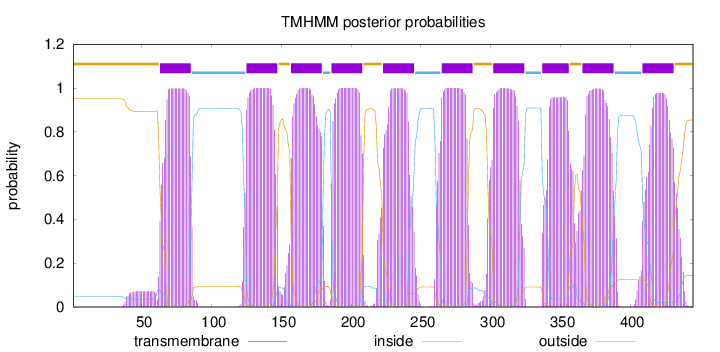
Topology
Length:
445
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
221.85282
Exp number, first 60 AAs:
1.52825
Total prob of N-in:
0.04856
outside
1 - 62
TMhelix
63 - 85
inside
86 - 124
TMhelix
125 - 147
outside
148 - 156
TMhelix
157 - 179
inside
180 - 185
TMhelix
186 - 208
outside
209 - 222
TMhelix
223 - 245
inside
246 - 264
TMhelix
265 - 287
outside
288 - 301
TMhelix
302 - 324
inside
325 - 336
TMhelix
337 - 356
outside
357 - 365
TMhelix
366 - 388
inside
389 - 408
TMhelix
409 - 431
outside
432 - 445
Population Genetic Test Statistics
Pi
13.424225
Theta
13.724101
Tajima's D
-0.843579
CLR
0.078847
CSRT
0.170241487925604
Interpretation
Uncertain
