Gene
KWMTBOMO00641
Annotation
PREDICTED:_protein_furry_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.483
Sequence
CDS
ATGCATTGTAATTACTACACGACGGACGGCGGTCGCAGTAGTCGCCGGGAAGGTCTGGGCTCCGCTGAAGCTACCGAAATGGTTTTGAATAACCTCTTCTACATAACAGCCAAGTTTTCGGACAATCACCCCAAAGAAATCGAAGAGCTGTGGTCGACGTTATGCGCCTGTTGGCCGAACAACCTAAAGGTCATCATCAGATATTTGATCATCGTCTCCGGAATGGCACCCAACGAACTTTTACCATATGCGAAACGCGTTGTCTTATATCTGGCCCGACCCCGGCCCGAGCGACTTCTTGATGAGATGATGACGGAACTGCAGACAGTCGAGACTTTGAACTGTCTCATCGAGCGCACGGAGACTCCGCCCTTCTACAGGCTCACATCCATGCGCAAGGCCACGTCGGGACACAGCGACGGACCCGGCACCGGCTCGCAGGACGCAACCGGTCGGACAGGTGAACTGGCGCCAGTCGAGAAAGGAACCATCCACACGAAGAGGCATAGCGGTGAAGATCCCGCCAAAGCAAGTGGAGCTAAGGCACCGGACACGCCCAACCCGAGACAGGACAAGCGGTCGTCAAACCCGCCGTGCTCAACGCCGCCCGCCCTGGAAGACGTTCCGCCGCCCTCTGATGGTGACGCCACCCCAACTGGCTTCGGTCCGGATTTAAGCGCTACGCCCCTTCAGGGCAGCGCTCCTTCGACGCCCCAGGAGGCGCGCGTGGACGTGCCCCAGCCGCGACCCCTCCCGATGCCGGAGTACGGCGGCTTCTTTGCTCTCCTCACCGAGTACTTGCCGGACAGCAACCAACCGATCTCCGGCTTCCACCGGTGCAACGTGGCGGTCATGCTGCTCTGTGACGTCGTCCTCGACGGAGTCGAGATAGACTGGTCTATTCATGTTCCGCTGATGCTACACATAGTGTTCCTCGGGATGGACCACACAAGGTGGATAGTTCACCAGCATTGCCGGCAGCTGCTACTGAACCTGCTGGTGGCGGTGGCCGCCCACCACGACCACCTGACGGTAGCGAAGACGCTGCTGGCGAGCCGGTCCGCTCAGCTGGGGCTCGGTACTCCGCCTACGCTGCCCATTGTTCAACACAACTTCACCGAGCCTGATGCGTCGTTTGATAAGCAGCCCCAATGCGCGCACTCGCCGAGGTCGCACGTCCGGTCCCTCAGCTCAGACCCTCAAACCACACCCGAAGGTGAAACCGATAACTTCCCGTCTCTCCCGGTGATCATAACCGGCGAGGCGGACGCGGACCCGATCCCGGAACACCCCGAGCCGGAGCCGGCCGAGATGCCGATCGCTGACGTCATCAAGTCATTGGTGCACTTCTTGGCGAATCGACCCACCCACATGCCGCTCTGGCAGTATGAGGACATTACCGCTAAAGTATGGACACTCCGGAGCTGTCAACAGATGCAGACCCTGGTGGGGCACGTGCTGCGAGTGTTCCGCGAGTCCCTGCCGCACGCGCTGGTGTCGGAGCGCTGGGCGCAGACCGCCCTGCAGCTCGGCCTGTCCTGTCCGTCGCGACACTACGCAGGACGATCCCTGCAGGTGTTCCGGTTCATCGGTGTAGCGATGACGGGCCGAATGGTCTCCGACATTCTCTCTCGTCTGGTGGAGACTGTTGCCGAGCAGGGAGAAGATATGCAGGGCTACGTCACGGAACTCCTTCTCACTCTGGAGGCGGCTGTTGATTCGCTCGAGTCAAACTTTAGGCCCCTGGACTACATGAGAGATATATTCAAATCGACGCCAAATTTGAATAATAAGGAGGGACAGCAACCTAACGGCGCTACGTACCCTGCAGTGATACCAGGCAAGCGTTCTCCCGGCGTGTGCGTGGGGTGCGTGAACAATCACACGAGGAGCACGAGCTACTCCGTGTCCTACTGCATCAGGAAGAGCAACGTGCAGGCTTCCGGATCTGACAAGCAGCACGAACGCGGCCGCGTGTGTCCTGAGGCCGGTAAAGGCGCCTGCTCCAACCTGTGCCGGTCGAGATCAGCGCAGTCACTGAAGCTGCTCGGCGACACCGCCACGCAGGATGATAAATTGACGATCCTCGCGACATTATTCTGGGTCGGAGCCTCCCTGTTGGAGAGCGACTATGAACACGAGTTCTTGCTAGGAGTCAGGTTGCTGTCCAGAGTCATGTGCCGGTTGCCGTTAGATAGACCCGACGCTAGAGAGAGGCTGGATCGCACTCAAGCTCACACATCCCCATCGCTGCATGCCCTGCTTCTAAAGGGCTGCACTCATCCCAACACGTACGAGCCTGTGGTCGGAGTACTAGCCGACATGATCCCACTACTGGAGCTGCCCGTCATCGATCCCACACAATCCGTGGCCTTCCCGATGACGGTAGTAGCTCTTCTGCCGTACATGTTGCTGCACTATGAAGACGCCAATCAGCTGTGCGTCAGAGCGGCCTGCCATATAGCACAGTTTACATCAGAGAAGAGTAAGAAACTAGAGAACTTGGGAACGGTGATGACTCTATACTCGCGTCGCACCTTCAGCAAGGAGAGCTTCCAGTGGACCAAGTGTGTCGTCAAATATCTGTGGGACACATATTCTCATCTCTCGCTGCAGATGCTGGCCTTTCTAGTGGAAGTACTCGAAAAGGGTTCGACCAGCCTGCAGCTACCGGTGCTCTCGATCCTGCACTGCATGCTGTACTACGTGGATCTGAACGCGGCGCCAGCTCAGCCGGTCAACACGGACCTGCTGCGAGCCGTCGCCAAGTTTATCGATCAAGAAGGAAACAACTACAGGGAGGCGATGAAGATATTGAAGTTGGTTGTGACCCGTTGCTCCACGCTGGTGGTGCCGCCGTCCTGGGAGAGCACCGCGTCCTCGCTGCTCGACGCCGAGATACACGGGAAGAAGGAGCTACCGGGACGAACCATGGATTTCACATTTGATCTGTCCCAAACGCCCGTAATCGGCAGGAAGTACCTACCGAAGGCCGGCAGTCAACCAGCCACCGGACCCAGCTCTCTATCAAATGCCTCCAGCACTACTCCAGACAAAGGCTCGTCAGCATCGCGCTCGGGCTGCGCGTCCACCGTGGTCGGTCCCGAGACCAGCACGTCACCGCGACGGTCGCTGTCACTGTCATTATCACCGGCCGACGCTCTGCTCTCCGGCTGGAAGAGACCTTGGATGTCTCAGAGTCGTGTTCGTGAATGCCTGGTGAACTTGCTAACGACATGCGGGCAACGCGTGGGTCTACCCAAGAGTCCTTCTGTGATATTCAGCCAAAGCTCGGAGTTGTTAGAGCGGGAATCTTCTATGGCGTCATCGCTTGAGGAGGTGTCTGGTACTCCTGGAAATGAGCCCTCAGGTGGAGCTCCCCCCACTGATCATTTTGGTGTCTTCAAGGATTTTGATTTTCTCGAATATGAAAGCGAGAGTATAGAGGGTGAAAGCTCAGATAATTTCAACTGGGGTGTCCGTCGGAGGCCACTAAGCGAAGAGCGAGATGAAGTCCAAATCCCTGAGAGGAGTCCTCCACCGCAGCAGCCGAGACCTCCTGAACTCCAGCAGATCACCAAACCTGCTGGCCATGATGACTCGTCCGATGAGGAAGCAGGGTGGGAGGAGGGGGCCACCCACGTGAGGAGCGAGGCAGACATGGCGACGCAACACGACATATCACTGAGGCCTAGAACACATTCGTTCTCAAGCGCCTCCAGCGGAGATCCTGACGGCGACCGCGGCGACGTGACCCCGGTGCCGGGCGCGGGCCCGGTGATGCGCGACGCGTGGCGCGCCTCCGTGGCCGCCCTCGTGCGCCACTCCGCGCACATGCAGCCCCTGGCCCTCATACACCGCCTGCACACCGTGCTCAAGGAAGTAACATCGAAGACGATCTCCCTGTGCTCGGACACGCCCTACACGAGCATCGGCTCGGCGGAGCTGTTCGCTGTACTGGAGAGGCTGAGCGCCGAAGAGCCGCCGCTCATCTGGGTGGGCCCCGGCGCCGCGGACTGCGCGAGGATCAGGGACGTCGTGCGCTACGCGTCCCTAGAGATCAACGAGCATCTCGAAACTTATTTCGATAGACGGGAACACGCTCTCGAGACGCTGGAGCTGCCGAGGAGTTCGAGTGAGGAGATGGGCCGATGCCTCTACAAGCTGGTGTTCCAACTCCTGCTGCTGCTGGAGACCAACAACAAGATCATTGTTGCCGTCTACCACGCCTCTCTGGACAATGGAGGCGTGGAGATGAGCGGTGCCGTGTGCGCGGTGCGTAACGCGCTGATCGCCGGCCACAGCGACTCCTGGGCCGAGCACACCCCCGGCCCGCACGACGACGCCAAGCTGTGGCAGCTCATGCAGTGGCAGAAATGGGGCGTTGCCCTGGCCCTCGTTCGGGAGCGCAACGCCGAGACGATCCTGGAGCGCGGAGAGATTCCCGAGGATGACGTCACCAGCCTGCTGCACGTCTACTGCAGGGGCTTAGCGCAGGCACATCCAGATATGGTCGCTATAACGCGGCCCGTGAACGATTTATCCCAGAATTTGTCCTTTATGATGGAGAGCCTGTACAAGGTGTCCGTGGCCCTCCAGCGGCAGGAGTTCGGCTGA
Protein
MHCNYYTTDGGRSSRREGLGSAEATEMVLNNLFYITAKFSDNHPKEIEELWSTLCACWPNNLKVIIRYLIIVSGMAPNELLPYAKRVVLYLARPRPERLLDEMMTELQTVETLNCLIERTETPPFYRLTSMRKATSGHSDGPGTGSQDATGRTGELAPVEKGTIHTKRHSGEDPAKASGAKAPDTPNPRQDKRSSNPPCSTPPALEDVPPPSDGDATPTGFGPDLSATPLQGSAPSTPQEARVDVPQPRPLPMPEYGGFFALLTEYLPDSNQPISGFHRCNVAVMLLCDVVLDGVEIDWSIHVPLMLHIVFLGMDHTRWIVHQHCRQLLLNLLVAVAAHHDHLTVAKTLLASRSAQLGLGTPPTLPIVQHNFTEPDASFDKQPQCAHSPRSHVRSLSSDPQTTPEGETDNFPSLPVIITGEADADPIPEHPEPEPAEMPIADVIKSLVHFLANRPTHMPLWQYEDITAKVWTLRSCQQMQTLVGHVLRVFRESLPHALVSERWAQTALQLGLSCPSRHYAGRSLQVFRFIGVAMTGRMVSDILSRLVETVAEQGEDMQGYVTELLLTLEAAVDSLESNFRPLDYMRDIFKSTPNLNNKEGQQPNGATYPAVIPGKRSPGVCVGCVNNHTRSTSYSVSYCIRKSNVQASGSDKQHERGRVCPEAGKGACSNLCRSRSAQSLKLLGDTATQDDKLTILATLFWVGASLLESDYEHEFLLGVRLLSRVMCRLPLDRPDARERLDRTQAHTSPSLHALLLKGCTHPNTYEPVVGVLADMIPLLELPVIDPTQSVAFPMTVVALLPYMLLHYEDANQLCVRAACHIAQFTSEKSKKLENLGTVMTLYSRRTFSKESFQWTKCVVKYLWDTYSHLSLQMLAFLVEVLEKGSTSLQLPVLSILHCMLYYVDLNAAPAQPVNTDLLRAVAKFIDQEGNNYREAMKILKLVVTRCSTLVVPPSWESTASSLLDAEIHGKKELPGRTMDFTFDLSQTPVIGRKYLPKAGSQPATGPSSLSNASSTTPDKGSSASRSGCASTVVGPETSTSPRRSLSLSLSPADALLSGWKRPWMSQSRVRECLVNLLTTCGQRVGLPKSPSVIFSQSSELLERESSMASSLEEVSGTPGNEPSGGAPPTDHFGVFKDFDFLEYESESIEGESSDNFNWGVRRRPLSEERDEVQIPERSPPPQQPRPPELQQITKPAGHDDSSDEEAGWEEGATHVRSEADMATQHDISLRPRTHSFSSASSGDPDGDRGDVTPVPGAGPVMRDAWRASVAALVRHSAHMQPLALIHRLHTVLKEVTSKTISLCSDTPYTSIGSAELFAVLERLSAEEPPLIWVGPGAADCARIRDVVRYASLEINEHLETYFDRREHALETLELPRSSSEEMGRCLYKLVFQLLLLLETNNKIIVAVYHASLDNGGVEMSGAVCAVRNALIAGHSDSWAEHTPGPHDDAKLWQLMQWQKWGVALALVRERNAETILERGEIPEDDVTSLLHVYCRGLAQAHPDMVAITRPVNDLSQNLSFMMESLYKVSVALQRQEFG
Summary
Uniprot
A0A2A4K139
A0A2A4JZF3
A0A212EKJ1
A0A2H1V2M2
A0A2W1BIQ0
A0A194PH62
+ More
A0A1B6DZ54 A0A1B6E7J2 A0A1B6BZF4 A0A1B6DEJ8 A0A151JY76 A0A1S4MZI1 A0A0P4VZM2 D6WD89 A0A023F4K0 E0VZU6 A0A1Y1NMT4 A0A1Y1NP32 A0A1W4XRH0 A0A0M8ZUN3 A0A146M8X4 V5GPS8 A0A0P6IV51 U4U4X5 N6TZ58 A0A182JYG3 A0A336KBU4 Q7PPU7 A0A084VU84 A0A1J1INL3 A0A336LX10 A0A336N1Q9 A0A1Q3G3U8 W5J544 A0A1Q3G3V5 A0A1Q3G3Y7 A0A182FE11 A0A2C9GWQ4 A0A2M4B8H5 A0A2M4A3D3 A0A182J7P8 A0A2C9GWQ8 A0A0J9RSF2 A0A0J9RS82 A0A0J9RSS1 A0A0J9RS87 A0A0R1DUT9 B4PEE3 A0A0R1E040 A0A0R1DUN4 A0A0Q5UIM0 B4LHR0 A0A0Q9WIE8 B3NCK0 A0A0Q5UFX9 A0A0Q5U3P7 A0A0Q5UAH1 A0A0Q9WTK4 A0A1W4U986 A0A1W4U8N2 A0A1W4U8N7 A0A1W4UKG4 A0A0J9RTT6 A0A0J9UI75 A0A0J9UI70 A0A0R1DUX4 A0A0R1DUK6 A0A0R1E0X3 A0A0R1DUM4 J9KAB8 A0A0R1DUL3 A0A0Q5UIH2 A0A0Q9WUD7 A0A0Q9WJE7 A0A0Q9WW00 A0A3B0K1S3 A0A3B0J612 A0A0Q5UIR5 A0A3B0JX91 A0A0Q5UET8 A0A0Q5UIV8 A0A0Q5U3Y8 A0A3B0JVD9 A0A0Q5U7K5 A0A0J9RTU3 A0A1W4UKJ9 A0A1W4UKK5 A0A0J9RSR6 A0A1W4U981 A0A1W4UMA9 A0A0J9RSF7 A0A0J9RSE7 A0A0J9RSQ9 A0A0J9UI64 A0A0J9RTT0 A0A0Q9WSD1 A0A0Q9WIZ1 A0A0Q9WUA2 A0A3B0JUF6
A0A1B6DZ54 A0A1B6E7J2 A0A1B6BZF4 A0A1B6DEJ8 A0A151JY76 A0A1S4MZI1 A0A0P4VZM2 D6WD89 A0A023F4K0 E0VZU6 A0A1Y1NMT4 A0A1Y1NP32 A0A1W4XRH0 A0A0M8ZUN3 A0A146M8X4 V5GPS8 A0A0P6IV51 U4U4X5 N6TZ58 A0A182JYG3 A0A336KBU4 Q7PPU7 A0A084VU84 A0A1J1INL3 A0A336LX10 A0A336N1Q9 A0A1Q3G3U8 W5J544 A0A1Q3G3V5 A0A1Q3G3Y7 A0A182FE11 A0A2C9GWQ4 A0A2M4B8H5 A0A2M4A3D3 A0A182J7P8 A0A2C9GWQ8 A0A0J9RSF2 A0A0J9RS82 A0A0J9RSS1 A0A0J9RS87 A0A0R1DUT9 B4PEE3 A0A0R1E040 A0A0R1DUN4 A0A0Q5UIM0 B4LHR0 A0A0Q9WIE8 B3NCK0 A0A0Q5UFX9 A0A0Q5U3P7 A0A0Q5UAH1 A0A0Q9WTK4 A0A1W4U986 A0A1W4U8N2 A0A1W4U8N7 A0A1W4UKG4 A0A0J9RTT6 A0A0J9UI75 A0A0J9UI70 A0A0R1DUX4 A0A0R1DUK6 A0A0R1E0X3 A0A0R1DUM4 J9KAB8 A0A0R1DUL3 A0A0Q5UIH2 A0A0Q9WUD7 A0A0Q9WJE7 A0A0Q9WW00 A0A3B0K1S3 A0A3B0J612 A0A0Q5UIR5 A0A3B0JX91 A0A0Q5UET8 A0A0Q5UIV8 A0A0Q5U3Y8 A0A3B0JVD9 A0A0Q5U7K5 A0A0J9RTU3 A0A1W4UKJ9 A0A1W4UKK5 A0A0J9RSR6 A0A1W4U981 A0A1W4UMA9 A0A0J9RSF7 A0A0J9RSE7 A0A0J9RSQ9 A0A0J9UI64 A0A0J9RTT0 A0A0Q9WSD1 A0A0Q9WIZ1 A0A0Q9WUA2 A0A3B0JUF6
Pubmed
EMBL
NWSH01000328
PCG77390.1
PCG77391.1
AGBW02014273
OWR42004.1
ODYU01000385
+ More
SOQ35081.1 KZ150031 PZC74738.1 KQ459605 KPI92049.1 GEDC01006325 JAS30973.1 GEDC01003419 JAS33879.1 GEDC01030928 JAS06370.1 GEDC01013273 JAS24025.1 KQ981515 KYN40775.1 AAZO01006539 GDKW01000562 JAI56033.1 KQ971321 EEZ99526.1 GBBI01002530 JAC16182.1 DS235854 EEB18902.1 GEZM01001083 JAV98175.1 GEZM01001082 JAV98176.1 KQ435835 KOX71500.1 GDHC01002740 JAQ15889.1 GALX01004874 JAB63592.1 GDUN01000319 JAN95600.1 KB631941 ERL87363.1 APGK01049762 KB741156 ENN73631.1 UFQS01000246 UFQT01000246 SSX01961.1 SSX22338.1 AAAB01008905 EAA09722.5 ATLV01016690 KE525103 KFB41528.1 CVRI01000054 CRL00073.1 SSX01963.1 SSX22340.1 UFQS01003345 UFQT01003345 SSX15527.1 SSX34893.1 GFDL01000568 JAV34477.1 ADMH02002184 ETN57970.1 GFDL01000565 JAV34480.1 GFDL01000566 JAV34479.1 GGFJ01000007 MBW49148.1 GGFK01001976 MBW35297.1 CM002912 KMY98703.1 KMY98696.1 KMY98707.1 KMY98701.1 CM000159 KRK00857.1 EDW92981.1 KRK00858.1 KRK00860.1 CH954178 KQS43719.1 CH940647 EDW69613.1 KRF84477.1 EDV51230.1 KQS43715.1 KQS43711.1 KQS43709.1 KRF84479.1 KMY98704.1 KMY98710.1 KMY98705.1 KRK00853.1 KRK00854.1 KRK00855.1 KRK00859.1 ABLF02029084 ABLF02029088 ABLF02029089 ABLF02029090 KRK00856.1 KQS43718.1 KRF84478.1 KRF84482.1 KRF84480.1 OUUW01000002 SPP77338.1 SPP77337.1 KQS43712.1 SPP77341.1 KQS43713.1 KQS43714.1 KQS43710.1 SPP77336.1 KQS43716.1 KMY98709.1 KMY98702.1 KMY98708.1 KMY98698.1 KMY98697.1 KMY98700.1 KMY98699.1 KRF84481.1 KRF84483.1 KRF84484.1 SPP77339.1
SOQ35081.1 KZ150031 PZC74738.1 KQ459605 KPI92049.1 GEDC01006325 JAS30973.1 GEDC01003419 JAS33879.1 GEDC01030928 JAS06370.1 GEDC01013273 JAS24025.1 KQ981515 KYN40775.1 AAZO01006539 GDKW01000562 JAI56033.1 KQ971321 EEZ99526.1 GBBI01002530 JAC16182.1 DS235854 EEB18902.1 GEZM01001083 JAV98175.1 GEZM01001082 JAV98176.1 KQ435835 KOX71500.1 GDHC01002740 JAQ15889.1 GALX01004874 JAB63592.1 GDUN01000319 JAN95600.1 KB631941 ERL87363.1 APGK01049762 KB741156 ENN73631.1 UFQS01000246 UFQT01000246 SSX01961.1 SSX22338.1 AAAB01008905 EAA09722.5 ATLV01016690 KE525103 KFB41528.1 CVRI01000054 CRL00073.1 SSX01963.1 SSX22340.1 UFQS01003345 UFQT01003345 SSX15527.1 SSX34893.1 GFDL01000568 JAV34477.1 ADMH02002184 ETN57970.1 GFDL01000565 JAV34480.1 GFDL01000566 JAV34479.1 GGFJ01000007 MBW49148.1 GGFK01001976 MBW35297.1 CM002912 KMY98703.1 KMY98696.1 KMY98707.1 KMY98701.1 CM000159 KRK00857.1 EDW92981.1 KRK00858.1 KRK00860.1 CH954178 KQS43719.1 CH940647 EDW69613.1 KRF84477.1 EDV51230.1 KQS43715.1 KQS43711.1 KQS43709.1 KRF84479.1 KMY98704.1 KMY98710.1 KMY98705.1 KRK00853.1 KRK00854.1 KRK00855.1 KRK00859.1 ABLF02029084 ABLF02029088 ABLF02029089 ABLF02029090 KRK00856.1 KQS43718.1 KRF84478.1 KRF84482.1 KRF84480.1 OUUW01000002 SPP77338.1 SPP77337.1 KQS43712.1 SPP77341.1 KQS43713.1 KQS43714.1 KQS43710.1 SPP77336.1 KQS43716.1 KMY98709.1 KMY98702.1 KMY98708.1 KMY98698.1 KMY98697.1 KMY98700.1 KMY98699.1 KRF84481.1 KRF84483.1 KRF84484.1 SPP77339.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K139
A0A2A4JZF3
A0A212EKJ1
A0A2H1V2M2
A0A2W1BIQ0
A0A194PH62
+ More
A0A1B6DZ54 A0A1B6E7J2 A0A1B6BZF4 A0A1B6DEJ8 A0A151JY76 A0A1S4MZI1 A0A0P4VZM2 D6WD89 A0A023F4K0 E0VZU6 A0A1Y1NMT4 A0A1Y1NP32 A0A1W4XRH0 A0A0M8ZUN3 A0A146M8X4 V5GPS8 A0A0P6IV51 U4U4X5 N6TZ58 A0A182JYG3 A0A336KBU4 Q7PPU7 A0A084VU84 A0A1J1INL3 A0A336LX10 A0A336N1Q9 A0A1Q3G3U8 W5J544 A0A1Q3G3V5 A0A1Q3G3Y7 A0A182FE11 A0A2C9GWQ4 A0A2M4B8H5 A0A2M4A3D3 A0A182J7P8 A0A2C9GWQ8 A0A0J9RSF2 A0A0J9RS82 A0A0J9RSS1 A0A0J9RS87 A0A0R1DUT9 B4PEE3 A0A0R1E040 A0A0R1DUN4 A0A0Q5UIM0 B4LHR0 A0A0Q9WIE8 B3NCK0 A0A0Q5UFX9 A0A0Q5U3P7 A0A0Q5UAH1 A0A0Q9WTK4 A0A1W4U986 A0A1W4U8N2 A0A1W4U8N7 A0A1W4UKG4 A0A0J9RTT6 A0A0J9UI75 A0A0J9UI70 A0A0R1DUX4 A0A0R1DUK6 A0A0R1E0X3 A0A0R1DUM4 J9KAB8 A0A0R1DUL3 A0A0Q5UIH2 A0A0Q9WUD7 A0A0Q9WJE7 A0A0Q9WW00 A0A3B0K1S3 A0A3B0J612 A0A0Q5UIR5 A0A3B0JX91 A0A0Q5UET8 A0A0Q5UIV8 A0A0Q5U3Y8 A0A3B0JVD9 A0A0Q5U7K5 A0A0J9RTU3 A0A1W4UKJ9 A0A1W4UKK5 A0A0J9RSR6 A0A1W4U981 A0A1W4UMA9 A0A0J9RSF7 A0A0J9RSE7 A0A0J9RSQ9 A0A0J9UI64 A0A0J9RTT0 A0A0Q9WSD1 A0A0Q9WIZ1 A0A0Q9WUA2 A0A3B0JUF6
A0A1B6DZ54 A0A1B6E7J2 A0A1B6BZF4 A0A1B6DEJ8 A0A151JY76 A0A1S4MZI1 A0A0P4VZM2 D6WD89 A0A023F4K0 E0VZU6 A0A1Y1NMT4 A0A1Y1NP32 A0A1W4XRH0 A0A0M8ZUN3 A0A146M8X4 V5GPS8 A0A0P6IV51 U4U4X5 N6TZ58 A0A182JYG3 A0A336KBU4 Q7PPU7 A0A084VU84 A0A1J1INL3 A0A336LX10 A0A336N1Q9 A0A1Q3G3U8 W5J544 A0A1Q3G3V5 A0A1Q3G3Y7 A0A182FE11 A0A2C9GWQ4 A0A2M4B8H5 A0A2M4A3D3 A0A182J7P8 A0A2C9GWQ8 A0A0J9RSF2 A0A0J9RS82 A0A0J9RSS1 A0A0J9RS87 A0A0R1DUT9 B4PEE3 A0A0R1E040 A0A0R1DUN4 A0A0Q5UIM0 B4LHR0 A0A0Q9WIE8 B3NCK0 A0A0Q5UFX9 A0A0Q5U3P7 A0A0Q5UAH1 A0A0Q9WTK4 A0A1W4U986 A0A1W4U8N2 A0A1W4U8N7 A0A1W4UKG4 A0A0J9RTT6 A0A0J9UI75 A0A0J9UI70 A0A0R1DUX4 A0A0R1DUK6 A0A0R1E0X3 A0A0R1DUM4 J9KAB8 A0A0R1DUL3 A0A0Q5UIH2 A0A0Q9WUD7 A0A0Q9WJE7 A0A0Q9WW00 A0A3B0K1S3 A0A3B0J612 A0A0Q5UIR5 A0A3B0JX91 A0A0Q5UET8 A0A0Q5UIV8 A0A0Q5U3Y8 A0A3B0JVD9 A0A0Q5U7K5 A0A0J9RTU3 A0A1W4UKJ9 A0A1W4UKK5 A0A0J9RSR6 A0A1W4U981 A0A1W4UMA9 A0A0J9RSF7 A0A0J9RSE7 A0A0J9RSQ9 A0A0J9UI64 A0A0J9RTT0 A0A0Q9WSD1 A0A0Q9WIZ1 A0A0Q9WUA2 A0A3B0JUF6
Ontologies
PANTHER
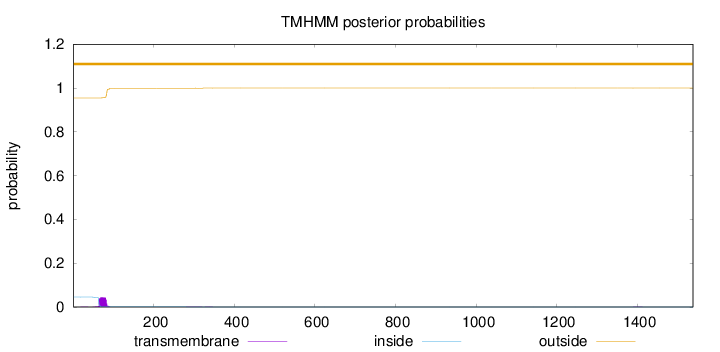
Topology
Length:
1538
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.922969999999997
Exp number, first 60 AAs:
0.02189
Total prob of N-in:
0.04555
outside
1 - 1538
Population Genetic Test Statistics
Pi
20.869272
Theta
18.275273
Tajima's D
0.451254
CLR
0.044912
CSRT
0.508974551272436
Interpretation
Uncertain
