Gene
KWMTBOMO00640
Pre Gene Modal
BGIBMGA003862
Annotation
PREDICTED:_H(+)/Cl(-)_exchange_transporter_5_isoform_X2_[Bombyx_mori]
Full name
Chloride channel protein
Location in the cell
PlasmaMembrane Reliability : 4.921
Sequence
CDS
ATGGAGAGGTTTCCTTTGAAGTCGTCAGTAACAAAAACAGGAATGACTTATCAATCAGTTTACAAAAAGGGCGCAAACACTGCGAATGGGCGTGGTGCTCATAGCGGTGAGGATGAAATGGTAGATATCACGCCGGGCCTCCAGCAAGCGGCGGACGGCGTCACCACACCCACAAGCCACGGAACCTTCCAGATCTACGAACAAGGAAGCCGAAATTCCGCGAGCAGCACTCATTCATACATTGCTCAGTTTTTCGGATCGTCGACAGGGGACACAATTATGTTCTCAGGAATGCAAGGTGATGATGATATTCCTGGAATTGGACAATATGAAGATTTCCATACTATTGATTGGCAAAGAGATATTGCGAGAGATCGCATGCGACATAGATATATTGTGAAGAAGAGGCAGGACTCAATATGGGATTTAATCAAAGGTGCCCATGATGCCTGGTCTGGATGGGTTTGTGTATTGCTTGTGGGAGTAGCAACTGGGATAGTGGCAGGAGTAATTGACATTGGTGCCTCATGGATGACAGATCTCAAATTTGGAATTTGTCCAGCTGCATTTTGGTTTAATCGAGAACAATGCTGCTGGTCAGACACAAATACTACATTCGACAGTGGGAATTGCTCACAGTGGCTCACCTGGCCTCAAGTTTTTGGCGACCGAGGTGATGGTGCTGGAGCTTACATCATCAGTTATCTCTTTTATATATTATGGGCTTTGATATTCGCTGCACTTTCAGCGTCGCTCGTAAGAATGTTCGCGCCCTATGCATGTGGCTCTGGTATCCCCGAAATTAAGACCATACTAAGTGGTTTCATTATCCGAGGCTACCTCGGCAAGTGGACCCTTATCATCAAAGTGGTAGGACTCATACTGTCTGTGTCATCGGGACTGTCTTTGGGGAAGGAAGGACCTATGGTGCACATTGCCAGCTGCCTTGGTAACATTCTATCATACCTGTTTCCGAAATACGGTCGCAACGAGGCCAAGAAGCGCGAGATCCTGTCCGCCGCCGCCGCCGCCGGAGTGTCCGTCGCATTCGGAGCTCCCATCGGTGGAGTACTCTTCAGTCTGGAAGAGGTATCGTACTATTTCCCGCTGAAGACCCTTTGGCGTTCGTTCTTCTGCGCTTTGATTGCCGCCTTCATACTGCGCTCAATCAATCCGTTCGGCAACGAGCACTCCGTGTTGTTCTTCGTCGAGTACAACAAGCCCTGGATCTTCTTCGAACTCATTCCGTTTGTTGGCCTCGGTATTATCGGGGGTTGCATCGCTACGATCTTCATAAAAGCGAATATCTATTGGTGTCGTTACCGGAAGTACTCGAAGCTGGGACAGTATCCGGTGACTGAAGTGCTGGTCGTGACGCTGATCACCGCCATCCTTGCCTACCCGAACCCATACACGCGCATGAACACCAGCGAACTCATATACCTGCTTTTTAACCAGTGCGGGATCTCTAACTCGGATCCTTTGTGTGACTACAATCGTAACTTCACCGATGCGAATTCGGCGATAGAGAAGGCGGCAGCCGGTGCTGGGGTGTACCGCGCTATCTGGCTGCTGGTACTCGCTCTGGTCTTCAAGCTGGTCGTCACCATCTTCACGTTCGGTATCAAGGTGCCGTGTGGACTGTTCATACCCAGCCTCGCGCTCGGAGCCATTGCTGGCAGGATTGTGGGCATCGGTGTCGAACAAATGGCATATACCTATCCGAAGATCTGGCTTTTCTCCGGTGAATGTTCAACCGGCGATGACTGTATAACTCCTGGACTATACGCGATGGTTGGAGCGGCGGCTGTACTCGGTGGAGTCACGCGGATGACCGTTTCGCTCGTTGTTATAATGTTCGAGTTGACCGGCGGGGTTCGTTACATCGTGCCCCTAATGGCGGCCGCCATGGCCTCCAAGTGGGTGGGAGACGCTTTGGGTAGACAAGGCATATATGACGCACACATCGCTCTGAACGGATATCCGTTTCTCGATAGCAAAGATGAATTCCAGCACACATCTCTCGCCGCCGACGTCATGCAACCTAAACGGAATGAAACTTTATCTGTAATAACTCAAGATTCGATGACCGTTGATGATGTGGAGACCCTGCTTAAGGAGACCGAACACAATGGGTATCCTGTTGTTGTGTCTAAGGAATCTCAGTACCTTGTTGGATTTGTGCTCCGACGGGATTTGAATCTTGCTATTGCAAACGCACGCCGCACCATCGAAGGCATCACTGGTCAATCTGTTGTGATCTTTGCCGGTAACGTTGCTCCTCCGCCGGGCCCGCCACCGTACCTCCAGCTCAGCCGCATCCTCGACATGGCTCCCATCACTGTCACCGATCAGACTCCCATGGAAACGGTTGTTGACATGTTCAGAAAATTGGGACTCCGCCAGACTCTGGTCACACATAACGGGCGGCTGCTCGGTGTCATCACTAAGAAGGACGTGCTCCGTCACGTGAAACATATGGACAACGAGGACCCGAGCTCAGTAATGTTTAACTGA
Protein
MERFPLKSSVTKTGMTYQSVYKKGANTANGRGAHSGEDEMVDITPGLQQAADGVTTPTSHGTFQIYEQGSRNSASSTHSYIAQFFGSSTGDTIMFSGMQGDDDIPGIGQYEDFHTIDWQRDIARDRMRHRYIVKKRQDSIWDLIKGAHDAWSGWVCVLLVGVATGIVAGVIDIGASWMTDLKFGICPAAFWFNREQCCWSDTNTTFDSGNCSQWLTWPQVFGDRGDGAGAYIISYLFYILWALIFAALSASLVRMFAPYACGSGIPEIKTILSGFIIRGYLGKWTLIIKVVGLILSVSSGLSLGKEGPMVHIASCLGNILSYLFPKYGRNEAKKREILSAAAAAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFCALIAAFILRSINPFGNEHSVLFFVEYNKPWIFFELIPFVGLGIIGGCIATIFIKANIYWCRYRKYSKLGQYPVTEVLVVTLITAILAYPNPYTRMNTSELIYLLFNQCGISNSDPLCDYNRNFTDANSAIEKAAAGAGVYRAIWLLVLALVFKLVVTIFTFGIKVPCGLFIPSLALGAIAGRIVGIGVEQMAYTYPKIWLFSGECSTGDDCITPGLYAMVGAAAVLGGVTRMTVSLVVIMFELTGGVRYIVPLMAAAMASKWVGDALGRQGIYDAHIALNGYPFLDSKDEFQHTSLAADVMQPKRNETLSVITQDSMTVDDVETLLKETEHNGYPVVVSKESQYLVGFVLRRDLNLAIANARRTIEGITGQSVVIFAGNVAPPPGPPPYLQLSRILDMAPITVTDQTPMETVVDMFRKLGLRQTLVTHNGRLLGVITKKDVLRHVKHMDNEDPSSVMFN
Summary
Similarity
Belongs to the chloride channel (TC 2.A.49) family.
Feature
chain Chloride channel protein
Uniprot
A0A2A4K0E0
A0A2A4JZE1
H9J2X5
A0A2H1VYK4
A0A2A4JZF9
A0A2A4K006
+ More
A0A1Y1K9R4 A0A194QXZ7 A0A336MTH3 A0A084WJP7 A0A182NJQ7 A0A182UT73 A0A182IKJ7 A0A3F2YS02 Q7Q6L5 Q17K86 A0A1S4EZP2 A0A3F2YS04 A0A182JRP5 A0A336MNN9 A0A182PHY9 A0A182QAB8 A0A182HWG9 A0A182SH22 A0A182RN01 A0A1Y1KA08 A0A2M4A7S7 A0A2M4A7U4 A0A1Y1K9N0 A0A1Y1KEZ1 W5J5H3 A0A182GME0 U5EIW3 V5GIB8 A0A1L8DF40 A0A182Y252 A0A2M4AAA1 A0A2J7PW34 A0A1W4WN47 A0A2J7PW41 A0A1L8DEW6 A0A067RLM4 A0A2J7PW37 A0A2A3E7Q1 A0A2J7PW36 B0WCM4 A0A088A776 A0A1W4WYZ0 A0A1W4WN61 A0A0C9S005 E2BWU6 A0A182L374 A0A195CVH7 A0A1Q3FEP5 A0A026WIM1 A0A3L8DFY2 A0A151X2A9 A0A154PC07 A0A1Q3FEN8 A0A0C9RB63 A0A182MS92 A0A1L8DDV0 A0A0C9RKD1 N6U4L6 D6WEX6 A0A0M8ZQ22 A0A1J1IEZ1 A0A182FHU9 A0A1Y0AWN2 E2A7M7 T1PCG6 F4WZJ2 A0A158NHA1 A0A0L0BQ37 A0A195DRD7 A0A195ET15 A0A195BXU1 A0A1I8PWN6 A0A0A1WZ08 A0A1I8PWS3 A0A034VPS9 A0A034VRM4 A0A0A1WIT1 A0A2J7PW40 A0A1B0B6Z7 B4GQU3 A0A0Q9WJ86 A0A1B0G3B8 A0A1A9YIC1 B4L0Z1 A0A3B0KED0 B3NDR6 Q2M106 B3M627 A0A0Q9XBT3 B4IU51 B4LGQ8 A0A0Q9XBT6 C9QP60 A0A0A9VY74
A0A1Y1K9R4 A0A194QXZ7 A0A336MTH3 A0A084WJP7 A0A182NJQ7 A0A182UT73 A0A182IKJ7 A0A3F2YS02 Q7Q6L5 Q17K86 A0A1S4EZP2 A0A3F2YS04 A0A182JRP5 A0A336MNN9 A0A182PHY9 A0A182QAB8 A0A182HWG9 A0A182SH22 A0A182RN01 A0A1Y1KA08 A0A2M4A7S7 A0A2M4A7U4 A0A1Y1K9N0 A0A1Y1KEZ1 W5J5H3 A0A182GME0 U5EIW3 V5GIB8 A0A1L8DF40 A0A182Y252 A0A2M4AAA1 A0A2J7PW34 A0A1W4WN47 A0A2J7PW41 A0A1L8DEW6 A0A067RLM4 A0A2J7PW37 A0A2A3E7Q1 A0A2J7PW36 B0WCM4 A0A088A776 A0A1W4WYZ0 A0A1W4WN61 A0A0C9S005 E2BWU6 A0A182L374 A0A195CVH7 A0A1Q3FEP5 A0A026WIM1 A0A3L8DFY2 A0A151X2A9 A0A154PC07 A0A1Q3FEN8 A0A0C9RB63 A0A182MS92 A0A1L8DDV0 A0A0C9RKD1 N6U4L6 D6WEX6 A0A0M8ZQ22 A0A1J1IEZ1 A0A182FHU9 A0A1Y0AWN2 E2A7M7 T1PCG6 F4WZJ2 A0A158NHA1 A0A0L0BQ37 A0A195DRD7 A0A195ET15 A0A195BXU1 A0A1I8PWN6 A0A0A1WZ08 A0A1I8PWS3 A0A034VPS9 A0A034VRM4 A0A0A1WIT1 A0A2J7PW40 A0A1B0B6Z7 B4GQU3 A0A0Q9WJ86 A0A1B0G3B8 A0A1A9YIC1 B4L0Z1 A0A3B0KED0 B3NDR6 Q2M106 B3M627 A0A0Q9XBT3 B4IU51 B4LGQ8 A0A0Q9XBT6 C9QP60 A0A0A9VY74
Pubmed
EMBL
NWSH01000328
PCG77388.1
PCG77385.1
BABH01031233
ODYU01005251
SOQ45919.1
+ More
PCG77387.1 PCG77389.1 GEZM01088448 JAV58193.1 KQ460953 KPJ10347.1 UFQT01000040 UFQT01001876 SSX18586.1 SSX32073.1 ATLV01024046 KE525348 KFB50441.1 APCN01001491 AAAB01008960 EAA11899.4 CH477227 EAT47099.1 SSX32074.1 AXCN02000515 GEZM01088447 GEZM01088445 JAV58194.1 GGFK01003522 MBW36843.1 GGFK01003536 MBW36857.1 GEZM01088449 JAV58192.1 GEZM01088446 JAV58195.1 ADMH02002165 ETN58115.1 JXUM01014075 JXUM01014076 JXUM01014077 KQ560393 KXJ82652.1 GANO01002472 JAB57399.1 GALX01004662 JAB63804.1 GFDF01009087 JAV04997.1 GGFK01004402 MBW37723.1 NEVH01020938 PNF20545.1 PNF20549.1 GFDF01009082 JAV05002.1 KK852550 KDR21510.1 PNF20547.1 KZ288353 PBC27312.1 PNF20548.1 DS231889 EDS43675.1 GBYB01013726 JAG83493.1 GL451188 EFN79810.1 KQ977259 KYN04542.1 GFDL01009027 JAV26018.1 KK107231 EZA54949.1 QOIP01000009 RLU19083.1 KQ982585 KYQ54386.1 KQ434869 KZC09353.1 GFDL01009029 JAV26016.1 GBYB01013724 JAG83491.1 AXCM01004096 GFDF01009529 JAV04555.1 GBYB01013727 JAG83494.1 APGK01043033 KB741011 ENN75556.1 KQ971318 EFA00320.2 KQ435916 KOX68790.1 CVRI01000047 CRK97566.1 KY921814 ART29403.1 GL437365 EFN70548.1 KA645633 AFP60262.1 GL888479 EGI60436.1 ADTU01001635 JRES01001546 KNC22121.1 KQ980612 KYN15069.1 KQ981979 KYN31373.1 KQ976398 KYM92761.1 GBXI01010411 JAD03881.1 GAKP01014473 GAKP01014471 JAC44479.1 GAKP01014472 JAC44480.1 GBXI01016019 JAC98272.1 PNF20546.1 JXJN01009324 CH479188 EDW40128.1 CH940647 KRF84956.1 CCAG010013759 CH933809 EDW18148.1 OUUW01000009 SPP84649.1 CH954178 EDV52199.2 CH379069 EAL30770.2 CH902618 EDV40743.2 KPU78741.1 KRG06000.1 CH891783 EDW99914.2 EDW70523.2 KRG05999.1 BT099942 ACX36518.1 GBHO01042472 GBHO01022630 GBHO01022625 GBHO01005800 JAG01132.1 JAG20974.1 JAG20979.1 JAG37804.1
PCG77387.1 PCG77389.1 GEZM01088448 JAV58193.1 KQ460953 KPJ10347.1 UFQT01000040 UFQT01001876 SSX18586.1 SSX32073.1 ATLV01024046 KE525348 KFB50441.1 APCN01001491 AAAB01008960 EAA11899.4 CH477227 EAT47099.1 SSX32074.1 AXCN02000515 GEZM01088447 GEZM01088445 JAV58194.1 GGFK01003522 MBW36843.1 GGFK01003536 MBW36857.1 GEZM01088449 JAV58192.1 GEZM01088446 JAV58195.1 ADMH02002165 ETN58115.1 JXUM01014075 JXUM01014076 JXUM01014077 KQ560393 KXJ82652.1 GANO01002472 JAB57399.1 GALX01004662 JAB63804.1 GFDF01009087 JAV04997.1 GGFK01004402 MBW37723.1 NEVH01020938 PNF20545.1 PNF20549.1 GFDF01009082 JAV05002.1 KK852550 KDR21510.1 PNF20547.1 KZ288353 PBC27312.1 PNF20548.1 DS231889 EDS43675.1 GBYB01013726 JAG83493.1 GL451188 EFN79810.1 KQ977259 KYN04542.1 GFDL01009027 JAV26018.1 KK107231 EZA54949.1 QOIP01000009 RLU19083.1 KQ982585 KYQ54386.1 KQ434869 KZC09353.1 GFDL01009029 JAV26016.1 GBYB01013724 JAG83491.1 AXCM01004096 GFDF01009529 JAV04555.1 GBYB01013727 JAG83494.1 APGK01043033 KB741011 ENN75556.1 KQ971318 EFA00320.2 KQ435916 KOX68790.1 CVRI01000047 CRK97566.1 KY921814 ART29403.1 GL437365 EFN70548.1 KA645633 AFP60262.1 GL888479 EGI60436.1 ADTU01001635 JRES01001546 KNC22121.1 KQ980612 KYN15069.1 KQ981979 KYN31373.1 KQ976398 KYM92761.1 GBXI01010411 JAD03881.1 GAKP01014473 GAKP01014471 JAC44479.1 GAKP01014472 JAC44480.1 GBXI01016019 JAC98272.1 PNF20546.1 JXJN01009324 CH479188 EDW40128.1 CH940647 KRF84956.1 CCAG010013759 CH933809 EDW18148.1 OUUW01000009 SPP84649.1 CH954178 EDV52199.2 CH379069 EAL30770.2 CH902618 EDV40743.2 KPU78741.1 KRG06000.1 CH891783 EDW99914.2 EDW70523.2 KRG05999.1 BT099942 ACX36518.1 GBHO01042472 GBHO01022630 GBHO01022625 GBHO01005800 JAG01132.1 JAG20974.1 JAG20979.1 JAG37804.1
Proteomes
UP000218220
UP000005204
UP000053240
UP000030765
UP000075884
UP000075903
+ More
UP000075880 UP000075840 UP000007062 UP000008820 UP000075881 UP000075885 UP000075886 UP000075901 UP000075900 UP000000673 UP000069940 UP000249989 UP000076408 UP000235965 UP000192223 UP000027135 UP000242457 UP000002320 UP000005203 UP000008237 UP000075882 UP000078542 UP000053097 UP000279307 UP000075809 UP000076502 UP000075883 UP000019118 UP000007266 UP000053105 UP000183832 UP000069272 UP000000311 UP000095301 UP000007755 UP000005205 UP000037069 UP000078492 UP000078541 UP000078540 UP000095300 UP000092460 UP000008744 UP000008792 UP000092444 UP000092443 UP000009192 UP000268350 UP000008711 UP000001819 UP000007801 UP000002282
UP000075880 UP000075840 UP000007062 UP000008820 UP000075881 UP000075885 UP000075886 UP000075901 UP000075900 UP000000673 UP000069940 UP000249989 UP000076408 UP000235965 UP000192223 UP000027135 UP000242457 UP000002320 UP000005203 UP000008237 UP000075882 UP000078542 UP000053097 UP000279307 UP000075809 UP000076502 UP000075883 UP000019118 UP000007266 UP000053105 UP000183832 UP000069272 UP000000311 UP000095301 UP000007755 UP000005205 UP000037069 UP000078492 UP000078541 UP000078540 UP000095300 UP000092460 UP000008744 UP000008792 UP000092444 UP000092443 UP000009192 UP000268350 UP000008711 UP000001819 UP000007801 UP000002282
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K0E0
A0A2A4JZE1
H9J2X5
A0A2H1VYK4
A0A2A4JZF9
A0A2A4K006
+ More
A0A1Y1K9R4 A0A194QXZ7 A0A336MTH3 A0A084WJP7 A0A182NJQ7 A0A182UT73 A0A182IKJ7 A0A3F2YS02 Q7Q6L5 Q17K86 A0A1S4EZP2 A0A3F2YS04 A0A182JRP5 A0A336MNN9 A0A182PHY9 A0A182QAB8 A0A182HWG9 A0A182SH22 A0A182RN01 A0A1Y1KA08 A0A2M4A7S7 A0A2M4A7U4 A0A1Y1K9N0 A0A1Y1KEZ1 W5J5H3 A0A182GME0 U5EIW3 V5GIB8 A0A1L8DF40 A0A182Y252 A0A2M4AAA1 A0A2J7PW34 A0A1W4WN47 A0A2J7PW41 A0A1L8DEW6 A0A067RLM4 A0A2J7PW37 A0A2A3E7Q1 A0A2J7PW36 B0WCM4 A0A088A776 A0A1W4WYZ0 A0A1W4WN61 A0A0C9S005 E2BWU6 A0A182L374 A0A195CVH7 A0A1Q3FEP5 A0A026WIM1 A0A3L8DFY2 A0A151X2A9 A0A154PC07 A0A1Q3FEN8 A0A0C9RB63 A0A182MS92 A0A1L8DDV0 A0A0C9RKD1 N6U4L6 D6WEX6 A0A0M8ZQ22 A0A1J1IEZ1 A0A182FHU9 A0A1Y0AWN2 E2A7M7 T1PCG6 F4WZJ2 A0A158NHA1 A0A0L0BQ37 A0A195DRD7 A0A195ET15 A0A195BXU1 A0A1I8PWN6 A0A0A1WZ08 A0A1I8PWS3 A0A034VPS9 A0A034VRM4 A0A0A1WIT1 A0A2J7PW40 A0A1B0B6Z7 B4GQU3 A0A0Q9WJ86 A0A1B0G3B8 A0A1A9YIC1 B4L0Z1 A0A3B0KED0 B3NDR6 Q2M106 B3M627 A0A0Q9XBT3 B4IU51 B4LGQ8 A0A0Q9XBT6 C9QP60 A0A0A9VY74
A0A1Y1K9R4 A0A194QXZ7 A0A336MTH3 A0A084WJP7 A0A182NJQ7 A0A182UT73 A0A182IKJ7 A0A3F2YS02 Q7Q6L5 Q17K86 A0A1S4EZP2 A0A3F2YS04 A0A182JRP5 A0A336MNN9 A0A182PHY9 A0A182QAB8 A0A182HWG9 A0A182SH22 A0A182RN01 A0A1Y1KA08 A0A2M4A7S7 A0A2M4A7U4 A0A1Y1K9N0 A0A1Y1KEZ1 W5J5H3 A0A182GME0 U5EIW3 V5GIB8 A0A1L8DF40 A0A182Y252 A0A2M4AAA1 A0A2J7PW34 A0A1W4WN47 A0A2J7PW41 A0A1L8DEW6 A0A067RLM4 A0A2J7PW37 A0A2A3E7Q1 A0A2J7PW36 B0WCM4 A0A088A776 A0A1W4WYZ0 A0A1W4WN61 A0A0C9S005 E2BWU6 A0A182L374 A0A195CVH7 A0A1Q3FEP5 A0A026WIM1 A0A3L8DFY2 A0A151X2A9 A0A154PC07 A0A1Q3FEN8 A0A0C9RB63 A0A182MS92 A0A1L8DDV0 A0A0C9RKD1 N6U4L6 D6WEX6 A0A0M8ZQ22 A0A1J1IEZ1 A0A182FHU9 A0A1Y0AWN2 E2A7M7 T1PCG6 F4WZJ2 A0A158NHA1 A0A0L0BQ37 A0A195DRD7 A0A195ET15 A0A195BXU1 A0A1I8PWN6 A0A0A1WZ08 A0A1I8PWS3 A0A034VPS9 A0A034VRM4 A0A0A1WIT1 A0A2J7PW40 A0A1B0B6Z7 B4GQU3 A0A0Q9WJ86 A0A1B0G3B8 A0A1A9YIC1 B4L0Z1 A0A3B0KED0 B3NDR6 Q2M106 B3M627 A0A0Q9XBT3 B4IU51 B4LGQ8 A0A0Q9XBT6 C9QP60 A0A0A9VY74
PDB
2JA3
E-value=6.13519e-56,
Score=554
Ontologies
GO
Topology
Subcellular location
Membrane
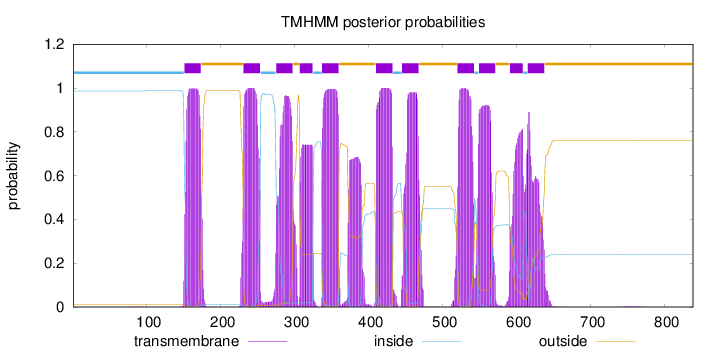
Length:
838
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
231.49545
Exp number, first 60 AAs:
0.00036
Total prob of N-in:
0.98702
inside
1 - 150
TMhelix
151 - 173
outside
174 - 230
TMhelix
231 - 253
inside
254 - 274
TMhelix
275 - 297
outside
298 - 306
TMhelix
307 - 324
inside
325 - 336
TMhelix
337 - 359
outside
360 - 409
TMhelix
410 - 432
inside
433 - 444
TMhelix
445 - 467
outside
468 - 519
TMhelix
520 - 542
inside
543 - 548
TMhelix
549 - 571
outside
572 - 590
TMhelix
591 - 608
inside
609 - 614
TMhelix
615 - 637
outside
638 - 838
Population Genetic Test Statistics
Pi
21.465294
Theta
19.804493
Tajima's D
0.219408
CLR
0.063803
CSRT
0.437578121093945
Interpretation
Uncertain
