Pre Gene Modal
BGIBMGA003894
Annotation
PREDICTED:_proton-coupled_amino_acid_transporter_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.932
Sequence
CDS
ATGACTGTCAAAGAAAAGATCGCCATTAGCCACATCAGCGGCAGCGCTGTCAGCCTCAGTCAAAATGGAAGCACCGTCACAGAGCCTTACGGAATGTTTCAATCTCGGGAGCAGATCCTGCCTCACACCAAGGGTCAGCTGGATATGGAGAGCGGAAAGACACGCGACGGACACAAAGTCAGGCATCCAACTTCATATTGGGACACACTACTCCACATCTTTAAAGGGAACATTAGTAGTGGTTTGTTGGCGATGGGAGACGCTTTCAAGAACGGAGGCATTGTATTCGCTCCTATAATGACAGCTATTTTAGGAGTCATCTGCGTACACGCACAACATTTATTGTTAAATGGTTCAGAAGAAATGCACAGACAAACTAAACAGGATCGGCCACCAGGGTTTGCCGAAACGGTGTACTTGGCGTTCGCGAAAGGTCCCCTCAGGCTCCGGCCCCTTGCCACCACCATGAAAATCGTCGTCAACTTCTTCCTGTGTATCACACAGCTCGGATTCTGCTGCGTTTATATTGTATTCATCGCTAACAATATTAAACTGATCTGCGACCATTATGAAGTGCACATCGACCTGTCGATTCACATGATTATAGTCGTGATTCCTGTCCTCCTGGCCTGCATGGTGCGAAACCTTAAGTACCTTACTCCGTTCTCTACAATCGCGAACATCATGATGGCAGTCGGAGTTAGCGTCGTTATTTACCAGGCGACCTTGGATTTACCATCGGTTCATACGAGGAATTACATAGCATCGTGGCAACAACTGCCTTTGTACTTCGGAACTGCTGTCTACGCTTTCGAAGGAATTGGTTTGGTGCTCCCCTTGAAGAACGAGATGAAGAATCCCGAAGAGTTTCAAAAGCCGTTCGGAGTGCTCAACATCGGAATGATCATAGTGGGCTCCATCTTCATTACCGTTGGATTCCTTGGTTACCTGAAGTGGGGTGAAGGAGTGGCGGGAAGTCTTACCTTGAACTTGGAGCCCGGTCACGTTTTGAGCAACGTAGTGCAAGTCTTGATAGGCTGTGCTGTTCTGTTCACGTATCCGCTTCAGTTCTACGTGCCCGTGGGCATCACCTGGCCGGCGCTGAAGAAGAGATGCGGTGAAAAATGTCTCATTGCCAAGGAGTTGGGCTATAGGGCTCTATTGGTGCTTTTGACTTTCATCCTGGCGGAATCCATACCCGAACTGGGCCTATTCATCTCCTTGGTTGGTGCTGTAAGCAGTACAGCTCTGGCACTCGTGTTCCCACCGCTGGTCGAGTTGGTCGTGGCCTCTCAGAAGAAAGAAGGACTTTCTGTCTTCATTATCGTAAAGGACACTATCATCATATTATTTGGATTATTCGTGTTCGTCACTGGTACATTTCAGAGTATCGCAAGTATTGTAAGAGCATTTAGAATTTAA
Protein
MTVKEKIAISHISGSAVSLSQNGSTVTEPYGMFQSREQILPHTKGQLDMESGKTRDGHKVRHPTSYWDTLLHIFKGNISSGLLAMGDAFKNGGIVFAPIMTAILGVICVHAQHLLLNGSEEMHRQTKQDRPPGFAETVYLAFAKGPLRLRPLATTMKIVVNFFLCITQLGFCCVYIVFIANNIKLICDHYEVHIDLSIHMIIVVIPVLLACMVRNLKYLTPFSTIANIMMAVGVSVVIYQATLDLPSVHTRNYIASWQQLPLYFGTAVYAFEGIGLVLPLKNEMKNPEEFQKPFGVLNIGMIIVGSIFITVGFLGYLKWGEGVAGSLTLNLEPGHVLSNVVQVLIGCAVLFTYPLQFYVPVGITWPALKKRCGEKCLIAKELGYRALLVLLTFILAESIPELGLFISLVGAVSSTALALVFPPLVELVVASQKKEGLSVFIIVKDTIIILFGLFVFVTGTFQSIASIVRAFRI
Summary
Uniprot
H9J307
A0A2A4ISW0
A0A2H1V866
A0A2W1B5V0
A0A212EQJ4
A0A0M8ZVB2
+ More
A0A026WKQ8 A0A195CUB5 A0A151J074 A0A0L7R999 A0A195FQF9 F4WMS2 A0A3L8DBN6 E2B4G0 A0A158NQ47 A0A154PJX2 A0A195BJ40 A0A310SH38 E2A1T3 N6UMC1 E2C8E6 A0A232F1W2 K7J624 K7J623 A0A139WKP6 A0A232F2K4 A0A1Q3FCE2 B0WJ42 Q16VP9 A0A1Q3FJL3 A0A1Q3FJJ1 B0WJ41 A0A182GXE1 A0A336MK74 A0A182GXE2 A0A1Y1N3G0 A0A2J7Q824 A0A2H8TZP0 J9K7G5 A0A2S2QED7 A0A182JDX4 A0A067RCN4 B4L4T5 A0A1B0DLT3 B4R6C2 A0A084VX27 Q9VX84 B3NWB5 Q29HX4 B4IEW9 B4NPC0 A0A1W4VJ46 B4Q255 A0A182KBU4 A0A088A4S3 Q5TV78 A0A1Y1MZY0 A0A182JIZ1 A0A3B0K2U8 A0A084VX26 W8BB83 A0A1Q3FJM6 A0A1B6CXX6 A0A1Q3FJI0 A0A034V4V8 T1PMG0 A0A0K8V1Y7 A0A2H8TYJ8 A0A034V2Q5 B4M2C5 A0A182N1B3 A0A336LBI0 A0A336M8V0 A0A1B6HWL4 A0A182IA96 A0A1L8DEA1 A0A1I8NCV9 A0A182XED2 A0A182GVY7 A0A182FIV1 A0A1B0C9X0 A0A0A1WFD2 A0A0A1XCL1 Q8MU61 A0A3Q0IZQ2 J9JIW5 A0A0L0C9Q7 A0A226E5W1 A0A0A1XDC3 A0A182LSC1 A0A1I8NXE2 A0A1B6M575 A0A2J7Q811 A0A1I8NXD6 A0A1B6JUV4 A0A2S2QIN8 A0A1D2NB39 A0A212FG41 A0A0P8ZEW7 A0A0P8Y3E4
A0A026WKQ8 A0A195CUB5 A0A151J074 A0A0L7R999 A0A195FQF9 F4WMS2 A0A3L8DBN6 E2B4G0 A0A158NQ47 A0A154PJX2 A0A195BJ40 A0A310SH38 E2A1T3 N6UMC1 E2C8E6 A0A232F1W2 K7J624 K7J623 A0A139WKP6 A0A232F2K4 A0A1Q3FCE2 B0WJ42 Q16VP9 A0A1Q3FJL3 A0A1Q3FJJ1 B0WJ41 A0A182GXE1 A0A336MK74 A0A182GXE2 A0A1Y1N3G0 A0A2J7Q824 A0A2H8TZP0 J9K7G5 A0A2S2QED7 A0A182JDX4 A0A067RCN4 B4L4T5 A0A1B0DLT3 B4R6C2 A0A084VX27 Q9VX84 B3NWB5 Q29HX4 B4IEW9 B4NPC0 A0A1W4VJ46 B4Q255 A0A182KBU4 A0A088A4S3 Q5TV78 A0A1Y1MZY0 A0A182JIZ1 A0A3B0K2U8 A0A084VX26 W8BB83 A0A1Q3FJM6 A0A1B6CXX6 A0A1Q3FJI0 A0A034V4V8 T1PMG0 A0A0K8V1Y7 A0A2H8TYJ8 A0A034V2Q5 B4M2C5 A0A182N1B3 A0A336LBI0 A0A336M8V0 A0A1B6HWL4 A0A182IA96 A0A1L8DEA1 A0A1I8NCV9 A0A182XED2 A0A182GVY7 A0A182FIV1 A0A1B0C9X0 A0A0A1WFD2 A0A0A1XCL1 Q8MU61 A0A3Q0IZQ2 J9JIW5 A0A0L0C9Q7 A0A226E5W1 A0A0A1XDC3 A0A182LSC1 A0A1I8NXE2 A0A1B6M575 A0A2J7Q811 A0A1I8NXD6 A0A1B6JUV4 A0A2S2QIN8 A0A1D2NB39 A0A212FG41 A0A0P8ZEW7 A0A0P8Y3E4
Pubmed
19121390
28756777
22118469
24508170
21719571
30249741
+ More
20798317 21347285 23537049 28648823 20075255 18362917 19820115 17510324 26483478 28004739 24845553 17994087 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 12364791 24495485 25348373 25315136 25830018 12609514 26108605 27289101 18057021
20798317 21347285 23537049 28648823 20075255 18362917 19820115 17510324 26483478 28004739 24845553 17994087 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 12364791 24495485 25348373 25315136 25830018 12609514 26108605 27289101 18057021
EMBL
BABH01031236
NWSH01007145
PCG63067.1
ODYU01001172
SOQ37007.1
KZ150373
+ More
PZC71128.1 AGBW02013268 OWR43772.1 KQ435828 KOX71865.1 KK107167 EZA56251.1 KQ977279 KYN04250.1 KQ980658 KYN14775.1 KQ414627 KOC67447.1 KQ981382 KYN42154.1 GL888218 EGI64549.1 QOIP01000010 RLU17741.1 GL445556 EFN89421.1 ADTU01022962 ADTU01022963 KQ434938 KZC12087.1 KQ976464 KYM84412.1 KQ769940 OAD52752.1 GL435798 EFN72605.1 APGK01017663 APGK01017664 APGK01017665 KB740047 ENN81841.1 GL453666 EFN75781.1 NNAY01001211 OXU24721.1 KQ971327 KYB28397.1 OXU24720.1 GFDL01009825 JAV25220.1 DS231955 EDS28921.1 CH477584 EAT38633.1 GFDL01007327 JAV27718.1 GFDL01007347 JAV27698.1 EDS28920.1 JXUM01095133 KQ564174 KXJ72624.1 UFQT01001380 SSX30295.1 JXUM01095134 KXJ72625.1 GEZM01016659 JAV91215.1 NEVH01016978 PNF24725.1 GFXV01007931 MBW19736.1 ABLF02031956 GGMS01006677 MBY75880.1 KK852550 KDR21512.1 CH933810 EDW07563.2 AJVK01006808 CM000366 EDX18172.1 ATLV01017839 ATLV01017840 KE525195 KFB42521.1 AE014298 BT133049 AAF48694.1 AEX93135.1 CH954180 EDV46454.2 CH379064 EAL31633.3 CH480832 EDW46223.1 CH964291 EDW86360.2 CM000162 EDX02563.1 AAAB01008846 EAL41169.4 GEZM01016660 JAV91214.1 OUUW01000015 SPP88587.1 ATLV01017837 KFB42520.1 GAMC01012277 JAB94278.1 GFDL01007225 JAV27820.1 GEDC01018972 JAS18326.1 GFDL01007264 JAV27781.1 GAKP01021398 GAKP01021393 JAC37559.1 KA649108 AFP63737.1 GDHF01019536 JAI32778.1 GFXV01006797 MBW18602.1 GAKP01021400 GAKP01021396 JAC37556.1 CH940651 EDW65829.2 UFQS01002451 UFQT01002451 SSX14073.1 SSX33491.1 UFQS01000552 UFQT01000552 SSX04840.1 SSX25203.1 GECU01028676 JAS79030.1 APCN01001135 GFDF01009369 JAV04715.1 JXUM01092012 KQ563958 KXJ73016.1 AJWK01002980 GBXI01017164 GBXI01010288 JAC97127.1 JAD04004.1 GBXI01005605 JAD08687.1 AJ457193 CAD29806.1 ABLF02023997 JRES01000720 KNC28986.1 LNIX01000006 OXA52690.1 GBXI01004908 JAD09384.1 AXCM01014760 GEBQ01008924 JAT31053.1 PNF24724.1 GECU01028613 GECU01004719 JAS79093.1 JAT02988.1 GGMS01008167 MBY77370.1 LJIJ01000111 ODN02463.1 AGBW02008720 OWR52705.1 CH902620 KPU73248.1 KPU73251.1 KPU73249.1
PZC71128.1 AGBW02013268 OWR43772.1 KQ435828 KOX71865.1 KK107167 EZA56251.1 KQ977279 KYN04250.1 KQ980658 KYN14775.1 KQ414627 KOC67447.1 KQ981382 KYN42154.1 GL888218 EGI64549.1 QOIP01000010 RLU17741.1 GL445556 EFN89421.1 ADTU01022962 ADTU01022963 KQ434938 KZC12087.1 KQ976464 KYM84412.1 KQ769940 OAD52752.1 GL435798 EFN72605.1 APGK01017663 APGK01017664 APGK01017665 KB740047 ENN81841.1 GL453666 EFN75781.1 NNAY01001211 OXU24721.1 KQ971327 KYB28397.1 OXU24720.1 GFDL01009825 JAV25220.1 DS231955 EDS28921.1 CH477584 EAT38633.1 GFDL01007327 JAV27718.1 GFDL01007347 JAV27698.1 EDS28920.1 JXUM01095133 KQ564174 KXJ72624.1 UFQT01001380 SSX30295.1 JXUM01095134 KXJ72625.1 GEZM01016659 JAV91215.1 NEVH01016978 PNF24725.1 GFXV01007931 MBW19736.1 ABLF02031956 GGMS01006677 MBY75880.1 KK852550 KDR21512.1 CH933810 EDW07563.2 AJVK01006808 CM000366 EDX18172.1 ATLV01017839 ATLV01017840 KE525195 KFB42521.1 AE014298 BT133049 AAF48694.1 AEX93135.1 CH954180 EDV46454.2 CH379064 EAL31633.3 CH480832 EDW46223.1 CH964291 EDW86360.2 CM000162 EDX02563.1 AAAB01008846 EAL41169.4 GEZM01016660 JAV91214.1 OUUW01000015 SPP88587.1 ATLV01017837 KFB42520.1 GAMC01012277 JAB94278.1 GFDL01007225 JAV27820.1 GEDC01018972 JAS18326.1 GFDL01007264 JAV27781.1 GAKP01021398 GAKP01021393 JAC37559.1 KA649108 AFP63737.1 GDHF01019536 JAI32778.1 GFXV01006797 MBW18602.1 GAKP01021400 GAKP01021396 JAC37556.1 CH940651 EDW65829.2 UFQS01002451 UFQT01002451 SSX14073.1 SSX33491.1 UFQS01000552 UFQT01000552 SSX04840.1 SSX25203.1 GECU01028676 JAS79030.1 APCN01001135 GFDF01009369 JAV04715.1 JXUM01092012 KQ563958 KXJ73016.1 AJWK01002980 GBXI01017164 GBXI01010288 JAC97127.1 JAD04004.1 GBXI01005605 JAD08687.1 AJ457193 CAD29806.1 ABLF02023997 JRES01000720 KNC28986.1 LNIX01000006 OXA52690.1 GBXI01004908 JAD09384.1 AXCM01014760 GEBQ01008924 JAT31053.1 PNF24724.1 GECU01028613 GECU01004719 JAS79093.1 JAT02988.1 GGMS01008167 MBY77370.1 LJIJ01000111 ODN02463.1 AGBW02008720 OWR52705.1 CH902620 KPU73248.1 KPU73251.1 KPU73249.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053105
UP000053097
UP000078542
+ More
UP000078492 UP000053825 UP000078541 UP000007755 UP000279307 UP000008237 UP000005205 UP000076502 UP000078540 UP000000311 UP000019118 UP000215335 UP000002358 UP000007266 UP000002320 UP000008820 UP000069940 UP000249989 UP000235965 UP000007819 UP000075880 UP000027135 UP000009192 UP000092462 UP000000304 UP000030765 UP000000803 UP000008711 UP000001819 UP000001292 UP000007798 UP000192221 UP000002282 UP000075881 UP000005203 UP000007062 UP000268350 UP000008792 UP000075884 UP000075840 UP000095301 UP000076407 UP000069272 UP000092461 UP000079169 UP000037069 UP000198287 UP000075883 UP000095300 UP000094527 UP000007801
UP000078492 UP000053825 UP000078541 UP000007755 UP000279307 UP000008237 UP000005205 UP000076502 UP000078540 UP000000311 UP000019118 UP000215335 UP000002358 UP000007266 UP000002320 UP000008820 UP000069940 UP000249989 UP000235965 UP000007819 UP000075880 UP000027135 UP000009192 UP000092462 UP000000304 UP000030765 UP000000803 UP000008711 UP000001819 UP000001292 UP000007798 UP000192221 UP000002282 UP000075881 UP000005203 UP000007062 UP000268350 UP000008792 UP000075884 UP000075840 UP000095301 UP000076407 UP000069272 UP000092461 UP000079169 UP000037069 UP000198287 UP000075883 UP000095300 UP000094527 UP000007801
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J307
A0A2A4ISW0
A0A2H1V866
A0A2W1B5V0
A0A212EQJ4
A0A0M8ZVB2
+ More
A0A026WKQ8 A0A195CUB5 A0A151J074 A0A0L7R999 A0A195FQF9 F4WMS2 A0A3L8DBN6 E2B4G0 A0A158NQ47 A0A154PJX2 A0A195BJ40 A0A310SH38 E2A1T3 N6UMC1 E2C8E6 A0A232F1W2 K7J624 K7J623 A0A139WKP6 A0A232F2K4 A0A1Q3FCE2 B0WJ42 Q16VP9 A0A1Q3FJL3 A0A1Q3FJJ1 B0WJ41 A0A182GXE1 A0A336MK74 A0A182GXE2 A0A1Y1N3G0 A0A2J7Q824 A0A2H8TZP0 J9K7G5 A0A2S2QED7 A0A182JDX4 A0A067RCN4 B4L4T5 A0A1B0DLT3 B4R6C2 A0A084VX27 Q9VX84 B3NWB5 Q29HX4 B4IEW9 B4NPC0 A0A1W4VJ46 B4Q255 A0A182KBU4 A0A088A4S3 Q5TV78 A0A1Y1MZY0 A0A182JIZ1 A0A3B0K2U8 A0A084VX26 W8BB83 A0A1Q3FJM6 A0A1B6CXX6 A0A1Q3FJI0 A0A034V4V8 T1PMG0 A0A0K8V1Y7 A0A2H8TYJ8 A0A034V2Q5 B4M2C5 A0A182N1B3 A0A336LBI0 A0A336M8V0 A0A1B6HWL4 A0A182IA96 A0A1L8DEA1 A0A1I8NCV9 A0A182XED2 A0A182GVY7 A0A182FIV1 A0A1B0C9X0 A0A0A1WFD2 A0A0A1XCL1 Q8MU61 A0A3Q0IZQ2 J9JIW5 A0A0L0C9Q7 A0A226E5W1 A0A0A1XDC3 A0A182LSC1 A0A1I8NXE2 A0A1B6M575 A0A2J7Q811 A0A1I8NXD6 A0A1B6JUV4 A0A2S2QIN8 A0A1D2NB39 A0A212FG41 A0A0P8ZEW7 A0A0P8Y3E4
A0A026WKQ8 A0A195CUB5 A0A151J074 A0A0L7R999 A0A195FQF9 F4WMS2 A0A3L8DBN6 E2B4G0 A0A158NQ47 A0A154PJX2 A0A195BJ40 A0A310SH38 E2A1T3 N6UMC1 E2C8E6 A0A232F1W2 K7J624 K7J623 A0A139WKP6 A0A232F2K4 A0A1Q3FCE2 B0WJ42 Q16VP9 A0A1Q3FJL3 A0A1Q3FJJ1 B0WJ41 A0A182GXE1 A0A336MK74 A0A182GXE2 A0A1Y1N3G0 A0A2J7Q824 A0A2H8TZP0 J9K7G5 A0A2S2QED7 A0A182JDX4 A0A067RCN4 B4L4T5 A0A1B0DLT3 B4R6C2 A0A084VX27 Q9VX84 B3NWB5 Q29HX4 B4IEW9 B4NPC0 A0A1W4VJ46 B4Q255 A0A182KBU4 A0A088A4S3 Q5TV78 A0A1Y1MZY0 A0A182JIZ1 A0A3B0K2U8 A0A084VX26 W8BB83 A0A1Q3FJM6 A0A1B6CXX6 A0A1Q3FJI0 A0A034V4V8 T1PMG0 A0A0K8V1Y7 A0A2H8TYJ8 A0A034V2Q5 B4M2C5 A0A182N1B3 A0A336LBI0 A0A336M8V0 A0A1B6HWL4 A0A182IA96 A0A1L8DEA1 A0A1I8NCV9 A0A182XED2 A0A182GVY7 A0A182FIV1 A0A1B0C9X0 A0A0A1WFD2 A0A0A1XCL1 Q8MU61 A0A3Q0IZQ2 J9JIW5 A0A0L0C9Q7 A0A226E5W1 A0A0A1XDC3 A0A182LSC1 A0A1I8NXE2 A0A1B6M575 A0A2J7Q811 A0A1I8NXD6 A0A1B6JUV4 A0A2S2QIN8 A0A1D2NB39 A0A212FG41 A0A0P8ZEW7 A0A0P8Y3E4
Ontologies
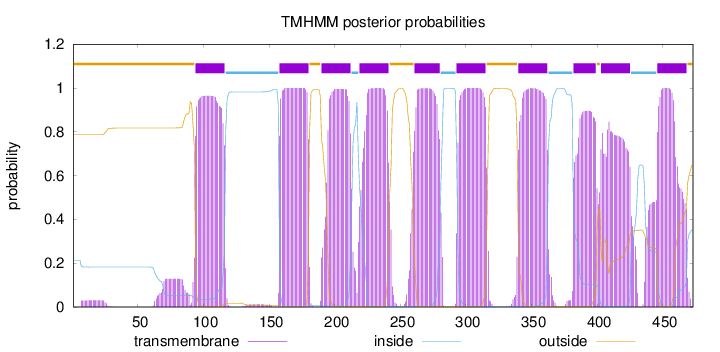
Topology
Length:
473
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
216.17758
Exp number, first 60 AAs:
0.59291
Total prob of N-in:
0.21262
outside
1 - 93
TMhelix
94 - 116
inside
117 - 157
TMhelix
158 - 180
outside
181 - 189
TMhelix
190 - 212
inside
213 - 218
TMhelix
219 - 241
outside
242 - 260
TMhelix
261 - 280
inside
281 - 292
TMhelix
293 - 315
outside
316 - 339
TMhelix
340 - 362
inside
363 - 381
TMhelix
382 - 399
outside
400 - 402
TMhelix
403 - 425
inside
426 - 445
TMhelix
446 - 468
outside
469 - 473
Population Genetic Test Statistics
Pi
15.014065
Theta
20.384771
Tajima's D
-1.5822
CLR
0.027917
CSRT
0.0497975101244938
Interpretation
Uncertain
