Gene
KWMTBOMO00635
Annotation
Retrovirus-related_Pol_polyprotein_from_transposon_TNT_1-94_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.798
Sequence
CDS
ATGTCGTCAAATAATACGATGGCACTGATTGAAAAGCTGACTGGAAGGGAAAACTATCCTACGTGGAGATTTGCAGTGCAAACATATCTCGAACACGAAGAATTATGGAGTTGTGTCACCGCGAAAGACAATGAAACAATTGATCCCAAACTTGATACAAAAGCTAAGTCAAAAATTATCTTATTAGTACATCCAATCAATTATGTACATATACAAGAAGCCAAAACAGCCAAGGAAGTTTGGAATAATTTAGCCAAAGCTTTCGATGACACGGGTTTGACACGCCGAGTGGGTCTGTTGAGAGATCTTATCAATACCACGCTTGAAAATTGTCTGAACATAGAAGATTATGTTAACAAGATTATGACCGCTGCCCATAAGTTAAGGAACATTGGTTTTAAAGTCGATGATGAATGGCTCGGGACTTTATTACTTGCTGGATTACCAGATATATATCAACCAATGATCATGGCACTAGAAAGTTCAGGTCTGACAATAACGGCTGATTCAGTGAAGACTAAGCTTCTACAAGATGTGAGAAACTCAGAGTCCAGTGCATTGTATGTCAGAAGCAAGGGGAATCAATTTGTCAAACAAACTCAAGATAAAACTAAAACTGGTAAAGGTCCAAGGTGCTATGTGTGTAATAAATATGGGCACATAAGTAAAAATTGCCGAAATAAGAAAAAGGAACAGAAGTCTAATGAGAACACAGATTCTGGTTATGTCGCTGTGTTATCTGCAACAATGAACAACGACAATAATTGGTACATTGATTCTGGAGCGTCAATGCATATGACAATGAATCGTGACTGGCTATATGATGAAATTACACCTCCAATTTCCACAATAAAAGTAGCTAATGACAAAAAGTTAGTTGTAAAAGCTTGTGGAAAGGTAAATCTTAATGTTGTCAATATGCAAGGCCAGTTAGAAACTATCAAGGTGCAGAATGTTCTCTATGTTCCTGAGTTGGCTACTAATCTTCTCTCTGTAAGTCAAATAATAAATAGTGGCTGTCAAGTACAATTTGACAAACAAGGATGTAAAATCTTAAATAAAACAAACAAGCAAGTTGCAGCTGCTAAAATGATAAATAATATGTACAGGCTAGTTACACACAGTGTTCCAGCATACACATCAGCTTCAACAGAGAATGATTCTTATTTATGGCATCAGAGAATGGCACATTTAAATTTTGAAGATTTAAATAAACTATCAGAAAGTGCAGGAGTCACATTAGAAAATACAGACAAAGTGATATGTATCTCATGTTTGGAAGGGAAGCAGACAAGATTTCCATTCCCTTCAGAAGGTTCCAGAGCACAAGCAAAGCTAGAACTTATTCACTCTGATGTCTGTGGTCCAATGGAGACACTGTCTATGGGAGGAGCACGCTACTTTCTTACATTCATCGATGATTTTACGAGGAAAGTATTTGTATATGTTTTAAAAAGAAGATCTGAAGTATTCCAAAAATTTCAAGAATTTAAAGTATTAGTTGAAAATCAACTTGATTTGACAATAAAAACTCTCCGCACAGATAATGGGGCTGAGTACTTGAGCAATGAATTTCAAGGTTTTCTTAAGAAATCTGGAATTATACATCAAACCTCCACTCCGTACTCGCCACAACAAAATGGTTTAGCGGAAAGAATGAACCGCACATTATTAGAAAGAGCCAGATGTATGCTTTTAAATGCAAAACTTCAGAAGCATTATTGGGCAGAAGCTGTGTCAACTGCAGCTTACATCACCAATAGATGTCCAACAAGAGCTTTAGCCTTTTTAACACCGGAAGAAATGTGGAGTGGTAAAAAGCCAGATATCACACATATGAAAATTTTTGGCTGTGAAGTGATGGTACATGTTCCTAAAGAAAAAGCTAAGAAGTGGGACTCAAAGACAAATAAATTGATATTCATTGGATATGGTGAACAAACCAAAGGATACAGGCTTTTAGATCCCAAAACAAGAAAAATCATTATAAGCAGAGATGTGATTTTTCTAGAAAGCTCAGTAAAAAGAAATTATGCTGTGGTACCTTTATCAGCATCAGTCCCAAATACCTGTGAAGATACTTCTGTGACTGAAAGTGGCACAACTTTACAAAATGTATCTGAAAGCATTGAAGGTATTAAGGATGAATCCAGTGATGATTTTTACTCAGATGAAGGATCTTTGTATGAACCAGAGATAAATATAGATCACCAGGAAGTAATGTCAAATGTAACTACAAGATCTAAAGCAAGACTGTGTCAAATGCAAGGAAATACCTATATGTGTAGAAAAGAAATTGTAGGACAGGAAAATATCCCAGAAACATATGATGAAGCTATATCTATTGAAAATGCTAATGCTGCAAAATGGAAACAATCAATTGAGGAAGAACTACAAGCACATGCAAAGAATAAAACTTGGCAATTAATTATGAAACCTGAAGGAGTGCGAGTGATTGGCTGTAAATGGGTCTTCAGAATAAAAGATGACCCATCGGGTCCGTGTTTTAAGTCAAGGCTATGTGCTAAAGGCTGTAGCCAGAAACAAGGAATTGATTATAAAGAGATATATTCTCCTACAGTGAGGTATGATTCAGTTCGAGTTTTACTTTCAGAAGCAGCACAACATAATTTGAATATAATTCAGTTTGATGTCAAAACTGCATTCTTATATGGTGACGTCAAAGAAGACATATATATGTTACCGCCTGAAGGTTTGGATACTAATGCAAATATGTAA
Protein
MSSNNTMALIEKLTGRENYPTWRFAVQTYLEHEELWSCVTAKDNETIDPKLDTKAKSKIILLVHPINYVHIQEAKTAKEVWNNLAKAFDDTGLTRRVGLLRDLINTTLENCLNIEDYVNKIMTAAHKLRNIGFKVDDEWLGTLLLAGLPDIYQPMIMALESSGLTITADSVKTKLLQDVRNSESSALYVRSKGNQFVKQTQDKTKTGKGPRCYVCNKYGHISKNCRNKKKEQKSNENTDSGYVAVLSATMNNDNNWYIDSGASMHMTMNRDWLYDEITPPISTIKVANDKKLVVKACGKVNLNVVNMQGQLETIKVQNVLYVPELATNLLSVSQIINSGCQVQFDKQGCKILNKTNKQVAAAKMINNMYRLVTHSVPAYTSASTENDSYLWHQRMAHLNFEDLNKLSESAGVTLENTDKVICISCLEGKQTRFPFPSEGSRAQAKLELIHSDVCGPMETLSMGGARYFLTFIDDFTRKVFVYVLKRRSEVFQKFQEFKVLVENQLDLTIKTLRTDNGAEYLSNEFQGFLKKSGIIHQTSTPYSPQQNGLAERMNRTLLERARCMLLNAKLQKHYWAEAVSTAAYITNRCPTRALAFLTPEEMWSGKKPDITHMKIFGCEVMVHVPKEKAKKWDSKTNKLIFIGYGEQTKGYRLLDPKTRKIIISRDVIFLESSVKRNYAVVPLSASVPNTCEDTSVTESGTTLQNVSESIEGIKDESSDDFYSDEGSLYEPEINIDHQEVMSNVTTRSKARLCQMQGNTYMCRKEIVGQENIPETYDEAISIENANAAKWKQSIEEELQAHAKNKTWQLIMKPEGVRVIGCKWVFRIKDDPSGPCFKSRLCAKGCSQKQGIDYKEIYSPTVRYDSVRVLLSEAAQHNLNIIQFDVKTAFLYGDVKEDIYMLPPEGLDTNANM
Summary
Uniprot
A0A194RU38
A0A2A4JRI1
A0A2A4J959
A0A194RJV9
A0A0J7NHJ8
A0A1Y1N668
+ More
A0A1Y1N932 A0A0A1WXQ3 A0A224XA26 A0A1W7R6K6 A0A1W7R6J6 A0A0N1ICY1 A0A0A9WPH5 A0A0A9WRE3 A0A1Y1LWI3 A0A1B6E9D0 E5SB91 A0A034VP70 A0A0V1APS7 A0A0V1FKH2 A0A0V1K1Y9 A0A0V0TUP0 A0A0V0T3I0 A0A0V1F2E6 A0A085MVP5 A0A1Y1N6L8 A0A085N572 A0A2J7R8S8 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R668 A0A0N1IFR1 A0A182GZK6 A0A0A9XEY5 A0A1Y3E9R1 A0A0V0Z6Q0 A0A0J7KG03 A0A0A9XVJ9 A0A1W7R657 A0A1Y1NAZ7 A0A1W7R6H6 A0A0A9WQD4 V5GAG2 A0A0A1XLZ7 A0A0V0WR33 A0A1Y1LYU4 W4VRQ4 A0A1W7R670 A0A1W7R663 A0A251T2S6 A0A1Y1N664 A0A251TN47 W4VRP9 A0A1Y1N3X2 W4VRP2 A0A0P6IXK4 W8ANX7 A0A251TCA0 A0A355BEE2 A0A2K3MYT4 A0A0J7K7B7 A0A0K8U956 A0A0V0YNF4 B6V6Z8 A0A0A9YFM0 B8YLY5 A0A0V0RWJ3 A0A182GZN1 W8BWJ0 B8YLY6 B8YLY3 A0A151TGQ2 B8YLY4 A0A151SDY6 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A151RJN4 K7JBY9 A0A2M3YZL3 T1PKV2 A0A146LFV8 A0A2M3YZL7 A0A1W7R6H7 A0A2M4CV35 W8AEF0 A0A151R6U8 A0A2N9HSV3 A0A1X7U495 W8ADM5 A0A2N9F1W0 A0A151R2G6 A0A2N9EE40 A0A2N9GHB5 A0A151TKT1 A0A0A1X768 A0A224X581 W8AL23
A0A1Y1N932 A0A0A1WXQ3 A0A224XA26 A0A1W7R6K6 A0A1W7R6J6 A0A0N1ICY1 A0A0A9WPH5 A0A0A9WRE3 A0A1Y1LWI3 A0A1B6E9D0 E5SB91 A0A034VP70 A0A0V1APS7 A0A0V1FKH2 A0A0V1K1Y9 A0A0V0TUP0 A0A0V0T3I0 A0A0V1F2E6 A0A085MVP5 A0A1Y1N6L8 A0A085N572 A0A2J7R8S8 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R668 A0A0N1IFR1 A0A182GZK6 A0A0A9XEY5 A0A1Y3E9R1 A0A0V0Z6Q0 A0A0J7KG03 A0A0A9XVJ9 A0A1W7R657 A0A1Y1NAZ7 A0A1W7R6H6 A0A0A9WQD4 V5GAG2 A0A0A1XLZ7 A0A0V0WR33 A0A1Y1LYU4 W4VRQ4 A0A1W7R670 A0A1W7R663 A0A251T2S6 A0A1Y1N664 A0A251TN47 W4VRP9 A0A1Y1N3X2 W4VRP2 A0A0P6IXK4 W8ANX7 A0A251TCA0 A0A355BEE2 A0A2K3MYT4 A0A0J7K7B7 A0A0K8U956 A0A0V0YNF4 B6V6Z8 A0A0A9YFM0 B8YLY5 A0A0V0RWJ3 A0A182GZN1 W8BWJ0 B8YLY6 B8YLY3 A0A151TGQ2 B8YLY4 A0A151SDY6 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A151RJN4 K7JBY9 A0A2M3YZL3 T1PKV2 A0A146LFV8 A0A2M3YZL7 A0A1W7R6H7 A0A2M4CV35 W8AEF0 A0A151R6U8 A0A2N9HSV3 A0A1X7U495 W8ADM5 A0A2N9F1W0 A0A151R2G6 A0A2N9EE40 A0A2N9GHB5 A0A151TKT1 A0A0A1X768 A0A224X581 W8AL23
Pubmed
EMBL
KQ459875
KPJ19636.1
NWSH01000816
PCG74073.1
NWSH01002292
PCG68657.1
+ More
PCG68658.1 KQ460297 KPJ16236.1 LBMM01004924 KMQ91985.1 GEZM01011856 JAV93393.1 GEZM01011857 JAV93390.1 GBXI01011094 JAD03198.1 GFTR01008662 JAW07764.1 GEHC01000867 JAV46778.1 GEHC01000868 JAV46777.1 KQ460882 KPJ11482.1 GBHO01033237 JAG10367.1 GBHO01033235 JAG10369.1 GEZM01045162 JAV77889.1 GEDC01002834 JAS34464.1 GAKP01015352 JAC43600.1 JYDH01000330 KRY26802.1 JYDT01000069 KRY86576.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 JYDJ01000136 KRX42725.1 JYDJ01000808 KRX33413.1 JYDR01000001 KRY80095.1 KL367628 KFD61291.1 GEZM01011858 GEZM01011854 JAV93389.1 KL367553 KFD64618.1 NEVH01006721 PNF37230.1 NEVH01019375 PNF22597.1 NEVH01002725 PNF41351.1 NEVH01006980 PNF36330.1 KQ460208 KPJ16606.1 JXUM01099713 KQ564513 KXJ72145.1 GBHO01026221 JAG17383.1 LVZM01023578 OUC39908.1 JYDQ01000379 KRY07987.1 LBMM01008124 KMQ89126.1 GBHO01020706 JAG22898.1 GEHC01000991 JAV46654.1 GEZM01011855 JAV93396.1 GEHC01000990 JAV46655.1 GBHO01036544 JAG07060.1 GALX01001351 JAB67115.1 GBXI01002679 JAD11613.1 JYDK01000071 KRX78146.1 GEZM01045163 JAV77888.1 GANO01000510 JAB59361.1 GEHC01000989 JAV46656.1 GEHC01000988 JAV46657.1 CM007901 OTG04806.1 GEZM01011859 JAV93384.1 CM007899 OTG12557.1 GANO01000659 JAB59212.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 GANO01000660 JAB59211.1 GDUN01000214 JAN95705.1 GAMC01020352 JAB86203.1 CM007900 OTG08534.1 DNYX01000353 HBK84078.1 ASHM01014005 PNX95963.1 LBMM01012669 KMQ86061.1 GDHF01029191 JAI23123.1 JYDU01000001 KRY01876.1 FJ238509 ACI62137.1 GBHO01011746 GBHO01011744 GBHO01011742 JAG31858.1 JAG31860.1 JAG31862.1 FJ544854 ACL97385.1 JYDL01000068 KRX18751.1 JXUM01099923 KQ564528 KXJ72133.1 GAMC01008929 JAB97626.1 FJ544856 ACL97386.1 FJ544851 ACL97383.1 CM003608 KYP66219.1 FJ544853 ACL97384.1 KQ483417 KYP53032.1 GBHO01024949 JAG18655.1 GBHO01024951 JAG18653.1 GBHO01024947 JAG18657.1 GBHO01024950 JAG18654.1 KQ483702 KYP42748.1 AAZX01000798 GGFM01000953 MBW21704.1 KA648745 AFP63374.1 GDHC01012180 JAQ06449.1 GGFM01000952 MBW21703.1 GEHC01000879 JAV46766.1 GGFL01004951 MBW69129.1 GAMC01019995 JAB86560.1 KQ484038 KYP38095.1 OIVN01003986 SPD14739.1 GAMC01020355 JAB86200.1 OIVN01000486 SPC81000.1 KQ484186 KYP36645.1 OIVN01000260 SPC77257.1 OIVN01001906 SPC98838.1 CM003607 KYP67664.1 GBXI01007143 JAD07149.1 GFTR01008771 JAW07655.1 GAMC01019998 JAB86557.1
PCG68658.1 KQ460297 KPJ16236.1 LBMM01004924 KMQ91985.1 GEZM01011856 JAV93393.1 GEZM01011857 JAV93390.1 GBXI01011094 JAD03198.1 GFTR01008662 JAW07764.1 GEHC01000867 JAV46778.1 GEHC01000868 JAV46777.1 KQ460882 KPJ11482.1 GBHO01033237 JAG10367.1 GBHO01033235 JAG10369.1 GEZM01045162 JAV77889.1 GEDC01002834 JAS34464.1 GAKP01015352 JAC43600.1 JYDH01000330 KRY26802.1 JYDT01000069 KRY86576.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 JYDJ01000136 KRX42725.1 JYDJ01000808 KRX33413.1 JYDR01000001 KRY80095.1 KL367628 KFD61291.1 GEZM01011858 GEZM01011854 JAV93389.1 KL367553 KFD64618.1 NEVH01006721 PNF37230.1 NEVH01019375 PNF22597.1 NEVH01002725 PNF41351.1 NEVH01006980 PNF36330.1 KQ460208 KPJ16606.1 JXUM01099713 KQ564513 KXJ72145.1 GBHO01026221 JAG17383.1 LVZM01023578 OUC39908.1 JYDQ01000379 KRY07987.1 LBMM01008124 KMQ89126.1 GBHO01020706 JAG22898.1 GEHC01000991 JAV46654.1 GEZM01011855 JAV93396.1 GEHC01000990 JAV46655.1 GBHO01036544 JAG07060.1 GALX01001351 JAB67115.1 GBXI01002679 JAD11613.1 JYDK01000071 KRX78146.1 GEZM01045163 JAV77888.1 GANO01000510 JAB59361.1 GEHC01000989 JAV46656.1 GEHC01000988 JAV46657.1 CM007901 OTG04806.1 GEZM01011859 JAV93384.1 CM007899 OTG12557.1 GANO01000659 JAB59212.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 GANO01000660 JAB59211.1 GDUN01000214 JAN95705.1 GAMC01020352 JAB86203.1 CM007900 OTG08534.1 DNYX01000353 HBK84078.1 ASHM01014005 PNX95963.1 LBMM01012669 KMQ86061.1 GDHF01029191 JAI23123.1 JYDU01000001 KRY01876.1 FJ238509 ACI62137.1 GBHO01011746 GBHO01011744 GBHO01011742 JAG31858.1 JAG31860.1 JAG31862.1 FJ544854 ACL97385.1 JYDL01000068 KRX18751.1 JXUM01099923 KQ564528 KXJ72133.1 GAMC01008929 JAB97626.1 FJ544856 ACL97386.1 FJ544851 ACL97383.1 CM003608 KYP66219.1 FJ544853 ACL97384.1 KQ483417 KYP53032.1 GBHO01024949 JAG18655.1 GBHO01024951 JAG18653.1 GBHO01024947 JAG18657.1 GBHO01024950 JAG18654.1 KQ483702 KYP42748.1 AAZX01000798 GGFM01000953 MBW21704.1 KA648745 AFP63374.1 GDHC01012180 JAQ06449.1 GGFM01000952 MBW21703.1 GEHC01000879 JAV46766.1 GGFL01004951 MBW69129.1 GAMC01019995 JAB86560.1 KQ484038 KYP38095.1 OIVN01003986 SPD14739.1 GAMC01020355 JAB86200.1 OIVN01000486 SPC81000.1 KQ484186 KYP36645.1 OIVN01000260 SPC77257.1 OIVN01001906 SPC98838.1 CM003607 KYP67664.1 GBXI01007143 JAD07149.1 GFTR01008771 JAW07655.1 GAMC01019998 JAB86557.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A194RU38
A0A2A4JRI1
A0A2A4J959
A0A194RJV9
A0A0J7NHJ8
A0A1Y1N668
+ More
A0A1Y1N932 A0A0A1WXQ3 A0A224XA26 A0A1W7R6K6 A0A1W7R6J6 A0A0N1ICY1 A0A0A9WPH5 A0A0A9WRE3 A0A1Y1LWI3 A0A1B6E9D0 E5SB91 A0A034VP70 A0A0V1APS7 A0A0V1FKH2 A0A0V1K1Y9 A0A0V0TUP0 A0A0V0T3I0 A0A0V1F2E6 A0A085MVP5 A0A1Y1N6L8 A0A085N572 A0A2J7R8S8 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R668 A0A0N1IFR1 A0A182GZK6 A0A0A9XEY5 A0A1Y3E9R1 A0A0V0Z6Q0 A0A0J7KG03 A0A0A9XVJ9 A0A1W7R657 A0A1Y1NAZ7 A0A1W7R6H6 A0A0A9WQD4 V5GAG2 A0A0A1XLZ7 A0A0V0WR33 A0A1Y1LYU4 W4VRQ4 A0A1W7R670 A0A1W7R663 A0A251T2S6 A0A1Y1N664 A0A251TN47 W4VRP9 A0A1Y1N3X2 W4VRP2 A0A0P6IXK4 W8ANX7 A0A251TCA0 A0A355BEE2 A0A2K3MYT4 A0A0J7K7B7 A0A0K8U956 A0A0V0YNF4 B6V6Z8 A0A0A9YFM0 B8YLY5 A0A0V0RWJ3 A0A182GZN1 W8BWJ0 B8YLY6 B8YLY3 A0A151TGQ2 B8YLY4 A0A151SDY6 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A151RJN4 K7JBY9 A0A2M3YZL3 T1PKV2 A0A146LFV8 A0A2M3YZL7 A0A1W7R6H7 A0A2M4CV35 W8AEF0 A0A151R6U8 A0A2N9HSV3 A0A1X7U495 W8ADM5 A0A2N9F1W0 A0A151R2G6 A0A2N9EE40 A0A2N9GHB5 A0A151TKT1 A0A0A1X768 A0A224X581 W8AL23
A0A1Y1N932 A0A0A1WXQ3 A0A224XA26 A0A1W7R6K6 A0A1W7R6J6 A0A0N1ICY1 A0A0A9WPH5 A0A0A9WRE3 A0A1Y1LWI3 A0A1B6E9D0 E5SB91 A0A034VP70 A0A0V1APS7 A0A0V1FKH2 A0A0V1K1Y9 A0A0V0TUP0 A0A0V0T3I0 A0A0V1F2E6 A0A085MVP5 A0A1Y1N6L8 A0A085N572 A0A2J7R8S8 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R668 A0A0N1IFR1 A0A182GZK6 A0A0A9XEY5 A0A1Y3E9R1 A0A0V0Z6Q0 A0A0J7KG03 A0A0A9XVJ9 A0A1W7R657 A0A1Y1NAZ7 A0A1W7R6H6 A0A0A9WQD4 V5GAG2 A0A0A1XLZ7 A0A0V0WR33 A0A1Y1LYU4 W4VRQ4 A0A1W7R670 A0A1W7R663 A0A251T2S6 A0A1Y1N664 A0A251TN47 W4VRP9 A0A1Y1N3X2 W4VRP2 A0A0P6IXK4 W8ANX7 A0A251TCA0 A0A355BEE2 A0A2K3MYT4 A0A0J7K7B7 A0A0K8U956 A0A0V0YNF4 B6V6Z8 A0A0A9YFM0 B8YLY5 A0A0V0RWJ3 A0A182GZN1 W8BWJ0 B8YLY6 B8YLY3 A0A151TGQ2 B8YLY4 A0A151SDY6 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A151RJN4 K7JBY9 A0A2M3YZL3 T1PKV2 A0A146LFV8 A0A2M3YZL7 A0A1W7R6H7 A0A2M4CV35 W8AEF0 A0A151R6U8 A0A2N9HSV3 A0A1X7U495 W8ADM5 A0A2N9F1W0 A0A151R2G6 A0A2N9EE40 A0A2N9GHB5 A0A151TKT1 A0A0A1X768 A0A224X581 W8AL23
PDB
5HRR
E-value=3.37809e-05,
Score=116
Ontologies
KEGG
GO
PANTHER
Topology
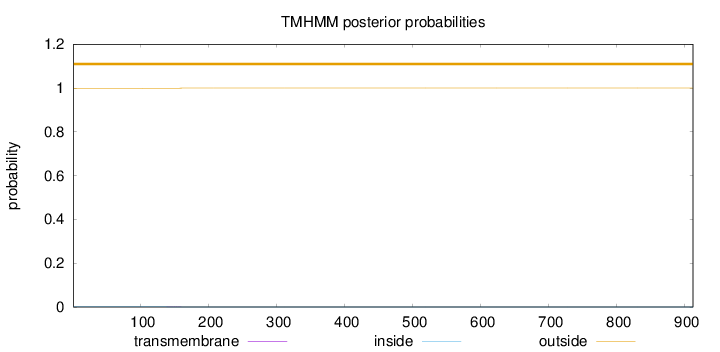
Length:
912
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01976
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.00103
outside
1 - 912
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
