Gene
KWMTBOMO00634
Annotation
Retrovirus-related_Pol_polyprotein_from_transposon_TNT_1-94_[Papilio_machaon]
Full name
Retrovirus-related Pol polyprotein from transposon TNT 1-94
Location in the cell
Nuclear Reliability : 3.453
Sequence
CDS
ATGTCGTCAAATAATACGATGGCACTGATTGAAAAGCTGACTGGAAGGGAAAACTATCCTACGTGGAGATTTGCAGTGCAAACATATCTCGAACACGAAGAATTATGGAGTTGTGTCACCGCGAAAGACAATGAAACAATTGATCCCAAACTTGATACAAAAGCTAAGTCAAAAATTATCTTATTAGTACATCCAATCAATTATGTACATATACAAGAAGCCAAAACAGCCAAGGAAGTTTGGAATAATTTAGCCAAAGCTTTCGATGACACGGGTTTGACACGCCGAGTGGGTCTGTTGAGAGATCTTATCAATACCACGCTTGAAAATTGTCTGAACATAGAAGATTATGTTAACAAGATTATGACCGCTGCCCATAAGTTAAGGAACATTGGTTTTAAAGTCGATGATGAATGGCTCGGGACTTTATTACTTGCTGGATTACCAGATATATATCAACCAATGATCATGGCACTAGAAAGTTCAGGTCTGACAATAACGGCTGATTCAGTGAAGACTAAGCTTCTACAAGATGTGAGAAACTCAGAGTCCAGTGCATTGTATGTCAGAAGCAAGGGGAATCAATTTGTCAAACAAACTCAAGATAAAACTAAAACTGGTAAAGGTCCAAGGTGCTATGTGTGTAATAAATATGGGCACATAAGTAAAAATTGCCGAAATAAGAAAAAGGAACAGAAGTCTAATGAGAACACAGATTCTGGTTATGTCGCTGTGTTATCTGCAACAATGAACAACGACAATAATTGGTACATTGATTCTGGAGCGTCAATGCATATGACAATGAATCGTGACTGGCTATATGATGAAATTACACCTCCAATTTCCACAATAAAAGTAGCTAATGACAAAAAGTTAGTTGTAAAAGCTTGTGGAAAGGTAAATCTTAATGTTGTCAATATGCAAGGCCAGTTAGAAACTATCAAGGTGCAGAATGTTCTCTATGTTCCTGAGTTGGCTACTAATCTTCTCTCTGTAAGTCAAATAATAAATAGTGGCTGTCAAGTACAATTTGACAAACAAGGATGTAAAATCTTAAATAAAACAAACAAGCAAGTTGCAGCTGCTAAAATGATAAATAATATGTACAGGCTAGTTACACACAGTGTTCCAGCATACACATCAGCTTCAACAGAGAATGATTCTTATTTATGGCATCAGAGAATGGCACATTTAAATTTTGAAGATTTAAATAAACTATCAGAAAGTGCAGGAGTCACATTAGAAAATACAGACAAAGTGATATGTATCTCATGTTTGGAAGGGAAGCAGACAAGATTTCCATTCCCTTCAGAAGGTTCCAGAGCACAAGCAAAGCTAGAACTTATTCACTCTGATGTCTGTGGTCCAATGGAGACACTGTCTATGGGAGGAGCACGCTACTTTCTTACATTCATCGATGATTTTACGAGGAAAGTATTTGTATATGTTTTAAAAAGAAGATCTGAAGTATTCCAAAAATTTCAAGAATTTAAAGTATTAGTTGAAAATCAACTTGATTTGACAATAAAAACTCTCCGCACAGATAATGGGGCTGAGTACTTGAGCAATGAATTTCAAGGTTTTCTTAAGAAATCTGGAATTATACATCAAACCTCCACTCCGTACTCGCCACAACAAAATGGTTTAGCGGAAAGAATGAACCGCACATTATTAGAAAGAGCCAGATGTATGCTTTTAAATGCAAAACTTCAGAAGCATTATTGGGCAGAAGCTGTGTCAACTGCAGCTTACATCACCAATAGATGTCCAACAAGAGCTTTAGCCTTTTTAACACCGGAAGAAATGTGGAGTGGTAAAAAGCCAGATATCACACATATGAAAATTTTTGGCTGTGAAGTGATGGTACATGTTCCTAAAGAAAAAGCTAAGAAGTGGGACTCAAAGACAAATAAATTGATATTCATTGGATATGGTGAACAAACCAAAGGATACAGGCTTTTAGATCCCAAAACAAGAAAAATCATTATAAGCAGAGATGTGATTTTTCTAGAAAGCTCAGTAAAAAGAAATTATGCTGTGGTACCTTTATCAGCATCAGTCCCAAATACCTGTGAAGATACTTCTGTGACTGAAAGTGGCACAACTTTACAAAATGTATCTGAAAGCATTGAAGGTATTAAGGATGAATCCAGTGATGATTTTTACTCAGATGAAGGATCTTTGTATGAACCAGAGATAAATATAGATCACCAGGAAGTAATGTCAAATGTAACTACAAGATCTAAAGCAAGACTGTGTCAAATGCAAGGAAATACCTATATGTGTAGAAAAGAAATTGTAGGACAGGAAAATATCCCAGAAACATATGATGAAGCTATATCTATTGAAAATGCTAATGCTGCAAAATGGAAACAATCAATTGAGGAAGAACTACAAGCACATGCAAAGAATAAAACTTGGCAATTAATTATGAAACCTGAAGGAGTGCGAGTGATTGGCTGTAAATGGGTCTTCAGAATAAAAGATGACCCATCGGGTCCGTGTTTTAAGTCAAGGCTATGTGCTAAAGGCTGTAGCCAGAAACAAGGAATTGATTATAAAGAGATATATTCTCCTACAGTGAGGTATGATTCAGTTCGAGTTTTACTTTCAGAAGCAGCACAACATAATTTGAATATAATTCAGTTTGATGTCAAAACTGCATTCTTATATGGTGACGTCAAAGAAGACATATATATGTTACCGCCTGAAGGTTTGGATACTAATGCAAATATGGTATGTAAACTCAATCGTTCTCTTTATGGCCTTAAACAAGCACCACGGTGTTGGAACCAAAAGTTTGACGCATCACTGAAGAAATTTGGTTTCAACAATAGTCATGCAGATAGGTGTTTGTATATTGGAAAAGTAAACCAGAACAAAGTTTATTTATTATTGTATGTTGATGATGGGCTTATACTTTCTAAGTCAAAGGATTCATTAAATACAGTAATGCAAGAATTAAAGCAAAATTTTGAAATAAAACAGTGTGATTCAAATAATTTTGTAGGATTACAAATTGAAAAATCTGAAAAATATGTCTTTATTCATCAGTCAAAATATATTGAGAGACTATTGTATAAATATAACATGCAAGATGCGAACTGCAATAATATCCCAGTAAGTCCACACACCATTCTGGAGAAAAGCACTGAGTATGTGGACAAGAATATTCCATACCGAGAGGCAGTTGGATCACTAATGCATCTGGCTGTGGTAAGCAGACCTGACATCATGTATGGAGTTAGTCTGGTAAGCAGATATCTTAATTGTTTTGATCACACTCACTGGAATGTAGTCAAGAAAATAATGAAATATTTAAAAGAAACTAAGGACTATGGCATTTGTTACACATCATCTCAGCAAAATAAAATTGAAGCTTATAGTGATGCTGATTTTGCTAAAGATAGTGCCACCCGAAAATCATTAACTGGGTATGTGTTCATTAAAAATGGAGCCGCCATAACATGGGCAACACAACGTCAGCAAAGTGTTGCTTTATCCACCACAGAGGCAGAATTTATGGCTGCTTGCTCTGCGACAAAAGAAGCTTTGTGGTTAAAGAGAATGTTATTAGATATTGGTGAATTTAATCAAGATTCTATATGCTTAAATATGGACAATCAAAGTGCAATCTGTTTAATTAAAAATGTGGACTATCACAAACGCTGTAAGCACATTGATGTTAAATATAATTTTATTAAAGAAAAATATAATGAAAAACATATAGATTTAAATTATGTTTGTACTAAAGAACAATATGCTGATATATTCACTAAAGCTCTGCCTAGGGATAAGTTTCAGTATCTTAGGAGTAAGCTTGGTATTAAGAAATTGAAATAG
Protein
MSSNNTMALIEKLTGRENYPTWRFAVQTYLEHEELWSCVTAKDNETIDPKLDTKAKSKIILLVHPINYVHIQEAKTAKEVWNNLAKAFDDTGLTRRVGLLRDLINTTLENCLNIEDYVNKIMTAAHKLRNIGFKVDDEWLGTLLLAGLPDIYQPMIMALESSGLTITADSVKTKLLQDVRNSESSALYVRSKGNQFVKQTQDKTKTGKGPRCYVCNKYGHISKNCRNKKKEQKSNENTDSGYVAVLSATMNNDNNWYIDSGASMHMTMNRDWLYDEITPPISTIKVANDKKLVVKACGKVNLNVVNMQGQLETIKVQNVLYVPELATNLLSVSQIINSGCQVQFDKQGCKILNKTNKQVAAAKMINNMYRLVTHSVPAYTSASTENDSYLWHQRMAHLNFEDLNKLSESAGVTLENTDKVICISCLEGKQTRFPFPSEGSRAQAKLELIHSDVCGPMETLSMGGARYFLTFIDDFTRKVFVYVLKRRSEVFQKFQEFKVLVENQLDLTIKTLRTDNGAEYLSNEFQGFLKKSGIIHQTSTPYSPQQNGLAERMNRTLLERARCMLLNAKLQKHYWAEAVSTAAYITNRCPTRALAFLTPEEMWSGKKPDITHMKIFGCEVMVHVPKEKAKKWDSKTNKLIFIGYGEQTKGYRLLDPKTRKIIISRDVIFLESSVKRNYAVVPLSASVPNTCEDTSVTESGTTLQNVSESIEGIKDESSDDFYSDEGSLYEPEINIDHQEVMSNVTTRSKARLCQMQGNTYMCRKEIVGQENIPETYDEAISIENANAAKWKQSIEEELQAHAKNKTWQLIMKPEGVRVIGCKWVFRIKDDPSGPCFKSRLCAKGCSQKQGIDYKEIYSPTVRYDSVRVLLSEAAQHNLNIIQFDVKTAFLYGDVKEDIYMLPPEGLDTNANMVCKLNRSLYGLKQAPRCWNQKFDASLKKFGFNNSHADRCLYIGKVNQNKVYLLLYVDDGLILSKSKDSLNTVMQELKQNFEIKQCDSNNFVGLQIEKSEKYVFIHQSKYIERLLYKYNMQDANCNNIPVSPHTILEKSTEYVDKNIPYREAVGSLMHLAVVSRPDIMYGVSLVSRYLNCFDHTHWNVVKKIMKYLKETKDYGICYTSSQQNKIEAYSDADFAKDSATRKSLTGYVFIKNGAAITWATQRQQSVALSTTEAEFMAACSATKEALWLKRMLLDIGEFNQDSICLNMDNQSAICLIKNVDYHKRCKHIDVKYNFIKEKYNEKHIDLNYVCTKEQYADIFTKALPRDKFQYLRSKLGIKKLK
Summary
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Keywords
Aspartyl protease
Complete proteome
Endonuclease
Hydrolase
Metal-binding
Nuclease
Nucleotidyltransferase
Protease
Reference proteome
RNA-directed DNA polymerase
Transferase
Transposable element
Zinc
Zinc-finger
Feature
chain Retrovirus-related Pol polyprotein from transposon TNT 1-94
Uniprot
A0A194RU38
A0A2A4JRI1
A0A2A4J959
A0A194RJV9
A0A0J7NHJ8
A0A0A1WXQ3
+ More
A0A1Y1N932 A0A1W7R6K6 A0A1W7R6J6 A0A0N1ICY1 E5SB91 A0A0V0TUP0 A0A0V1APS7 A0A0A9WRE3 A0A1Y1MYL8 A0A1Y1MYL6 A0A224XA26 A0A1Y1LYU4 A0A0J7K725 A0A0A9WQD4 A0A1B6E9D0 B8YLY5 A0A0V1FKH2 A0A0V1K1Y9 A0A0V1F2E6 A0A151SDY6 B8YLY6 A0A151RJN4 B8YLY3 B8YLY4 A0A151R6U8 A0A151R2G6 A0A151TKT1 H6U1F0 A0A0P6IXK4 A0A2N9FMR2 A0A2N9GLP2 A0A2N9IUS6 Q6BCY1 P10978 H6U1E9 B8YLY7 A0A1Y1N664 A0A0A1XLZ7 A0A2S2NXV8 A0A251TN47 A0A1Y1N3G5 T1PKV2 A0A2M3YZL3 W8BWJ0 A0A034VP70 A0A2M3YZL7 A0A2M4CV35 A0A151RCT9 K7J8U2 B1N668 A0A0V1B5K0 A0A0V1B5A2 A0A2N9GHK9 A0A251TCA0 A0A0V0YNF4 A0A1Y1M571 B6V6Z8 A0A2N9ETC8 A0A151THM1 A0A2N9GY85 E5SJ15 A0A2N9JB15 A0A0V0RWJ3 A0A2R6RYD4 E5SZD6 A0A2N9FQJ1 A0A1W7R6C8 A0A0J7KI47 A0A2K3NXK0 A0A182GSW5 A0A2N9HLU0 A0A139WFV5 A0A2N9H4V1 A0A1W7R6H7 A0A0V1M1K0 A0A2N9IWX1 A0A1Y1KNM9 A0A2P6SP23 A0A2N9F1W0 Q9ZRJ0 A0A2N9GHB5 A0A2N9FWL6 A0A2N9IGN3 A0A224X581 A0A0A9XD75 A0A0A9XFC8 A5B5N4 A0A0V1GS48 A0A0V0V098 A5AS37 A0A2N9EE21 A0A251T6N3 A0A0V1NJN8 A0A0A1X768
A0A1Y1N932 A0A1W7R6K6 A0A1W7R6J6 A0A0N1ICY1 E5SB91 A0A0V0TUP0 A0A0V1APS7 A0A0A9WRE3 A0A1Y1MYL8 A0A1Y1MYL6 A0A224XA26 A0A1Y1LYU4 A0A0J7K725 A0A0A9WQD4 A0A1B6E9D0 B8YLY5 A0A0V1FKH2 A0A0V1K1Y9 A0A0V1F2E6 A0A151SDY6 B8YLY6 A0A151RJN4 B8YLY3 B8YLY4 A0A151R6U8 A0A151R2G6 A0A151TKT1 H6U1F0 A0A0P6IXK4 A0A2N9FMR2 A0A2N9GLP2 A0A2N9IUS6 Q6BCY1 P10978 H6U1E9 B8YLY7 A0A1Y1N664 A0A0A1XLZ7 A0A2S2NXV8 A0A251TN47 A0A1Y1N3G5 T1PKV2 A0A2M3YZL3 W8BWJ0 A0A034VP70 A0A2M3YZL7 A0A2M4CV35 A0A151RCT9 K7J8U2 B1N668 A0A0V1B5K0 A0A0V1B5A2 A0A2N9GHK9 A0A251TCA0 A0A0V0YNF4 A0A1Y1M571 B6V6Z8 A0A2N9ETC8 A0A151THM1 A0A2N9GY85 E5SJ15 A0A2N9JB15 A0A0V0RWJ3 A0A2R6RYD4 E5SZD6 A0A2N9FQJ1 A0A1W7R6C8 A0A0J7KI47 A0A2K3NXK0 A0A182GSW5 A0A2N9HLU0 A0A139WFV5 A0A2N9H4V1 A0A1W7R6H7 A0A0V1M1K0 A0A2N9IWX1 A0A1Y1KNM9 A0A2P6SP23 A0A2N9F1W0 Q9ZRJ0 A0A2N9GHB5 A0A2N9FWL6 A0A2N9IGN3 A0A224X581 A0A0A9XD75 A0A0A9XFC8 A5B5N4 A0A0V1GS48 A0A0V0V098 A5AS37 A0A2N9EE21 A0A251T6N3 A0A0V1NJN8 A0A0A1X768
Pubmed
EMBL
KQ459875
KPJ19636.1
NWSH01000816
PCG74073.1
NWSH01002292
PCG68657.1
+ More
PCG68658.1 KQ460297 KPJ16236.1 LBMM01004924 KMQ91985.1 GBXI01011094 JAD03198.1 GEZM01011857 JAV93390.1 GEHC01000867 JAV46778.1 GEHC01000868 JAV46777.1 KQ460882 KPJ11482.1 JYDJ01000136 KRX42725.1 JYDH01000330 KRY26802.1 GBHO01033235 JAG10369.1 GEZM01017328 JAV90763.1 GEZM01017329 JAV90762.1 GFTR01008662 JAW07764.1 GEZM01045163 GEZM01045162 JAV77888.1 LBMM01012536 KMQ86167.1 GBHO01036544 JAG07060.1 GEDC01002834 JAS34464.1 FJ544854 ACL97385.1 JYDT01000069 KRY86576.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 JYDR01000001 KRY80095.1 KQ483417 KYP53032.1 FJ544856 ACL97386.1 KQ483702 KYP42748.1 FJ544851 ACL97383.1 FJ544853 ACL97384.1 KQ484038 KYP38095.1 KQ484186 KYP36645.1 CM003607 KYP67664.1 JN402333 AFB73912.1 GDUN01000214 JAN95705.1 OIVN01000991 OIVN01001835 SPC88370.1 SPC98114.1 OIVN01002055 SPD00174.1 OIVN01006261 SPD29147.1 AB162659 BAD34493.1 X13777 JN402332 AFB73911.1 FJ544857 ACL97387.1 GEZM01011859 JAV93384.1 GBXI01002679 JAD11613.1 GGMR01009345 MBY21964.1 CM007899 OTG12557.1 GEZM01013586 JAV92451.1 KA648745 AFP63374.1 GGFM01000953 MBW21704.1 GAMC01008929 JAB97626.1 GAKP01015352 JAC43600.1 GGFM01000952 MBW21703.1 GGFL01004951 MBW69129.1 KQ483843 KYP40337.1 AAZX01000499 EF094939 EF094940 ABO36622.1 ABO36636.1 JYDH01000102 KRY32233.1 KRY32232.1 OIVN01001931 SPC99063.1 CM007900 OTG08534.1 JYDU01000001 KRY01876.1 GEZM01040221 GEZM01040220 JAV80932.1 FJ238509 ACI62137.1 OIVN01000557 SPC82046.1 CM003608 KYP66486.1 OIVN01002516 SPD04274.1 OIVN01006466 SPD33659.1 JYDL01000068 KRX18751.1 NKQK01000002 PSS35035.1 OIVN01001052 SPC89229.1 GEHC01000927 JAV46718.1 LBMM01007046 KMQ90088.1 ASHM01002084 PNY07765.1 JXUM01085595 KQ563513 KXJ73717.1 OIVN01003683 SPD12935.1 KQ971352 KYB26731.1 OIVN01002854 SPD06945.1 GEHC01000879 JAV46766.1 JYDO01000312 KRZ65659.1 OIVN01006239 SPD28653.1 GEZM01078128 GEZM01078127 JAV63002.1 PDCK01000039 PRQ60431.1 OIVN01000486 SPC81000.1 D83003 BAA11674.1 OIVN01001906 SPC98838.1 OIVN01001534 SPC95017.1 OIVN01005591 SPD23209.1 GFTR01008771 JAW07655.1 GBHO01024947 JAG18657.1 GBHO01024950 JAG18654.1 AM447500 CAN74198.1 JYDP01000350 KRZ01021.1 JYDN01000113 KRX56900.1 AM433482 CAN69340.1 OIVN01000036 SPC73022.1 OTG06758.1 JYDM01000185 KRZ84108.1 GBXI01007143 JAD07149.1
PCG68658.1 KQ460297 KPJ16236.1 LBMM01004924 KMQ91985.1 GBXI01011094 JAD03198.1 GEZM01011857 JAV93390.1 GEHC01000867 JAV46778.1 GEHC01000868 JAV46777.1 KQ460882 KPJ11482.1 JYDJ01000136 KRX42725.1 JYDH01000330 KRY26802.1 GBHO01033235 JAG10369.1 GEZM01017328 JAV90763.1 GEZM01017329 JAV90762.1 GFTR01008662 JAW07764.1 GEZM01045163 GEZM01045162 JAV77888.1 LBMM01012536 KMQ86167.1 GBHO01036544 JAG07060.1 GEDC01002834 JAS34464.1 FJ544854 ACL97385.1 JYDT01000069 KRY86576.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 JYDR01000001 KRY80095.1 KQ483417 KYP53032.1 FJ544856 ACL97386.1 KQ483702 KYP42748.1 FJ544851 ACL97383.1 FJ544853 ACL97384.1 KQ484038 KYP38095.1 KQ484186 KYP36645.1 CM003607 KYP67664.1 JN402333 AFB73912.1 GDUN01000214 JAN95705.1 OIVN01000991 OIVN01001835 SPC88370.1 SPC98114.1 OIVN01002055 SPD00174.1 OIVN01006261 SPD29147.1 AB162659 BAD34493.1 X13777 JN402332 AFB73911.1 FJ544857 ACL97387.1 GEZM01011859 JAV93384.1 GBXI01002679 JAD11613.1 GGMR01009345 MBY21964.1 CM007899 OTG12557.1 GEZM01013586 JAV92451.1 KA648745 AFP63374.1 GGFM01000953 MBW21704.1 GAMC01008929 JAB97626.1 GAKP01015352 JAC43600.1 GGFM01000952 MBW21703.1 GGFL01004951 MBW69129.1 KQ483843 KYP40337.1 AAZX01000499 EF094939 EF094940 ABO36622.1 ABO36636.1 JYDH01000102 KRY32233.1 KRY32232.1 OIVN01001931 SPC99063.1 CM007900 OTG08534.1 JYDU01000001 KRY01876.1 GEZM01040221 GEZM01040220 JAV80932.1 FJ238509 ACI62137.1 OIVN01000557 SPC82046.1 CM003608 KYP66486.1 OIVN01002516 SPD04274.1 OIVN01006466 SPD33659.1 JYDL01000068 KRX18751.1 NKQK01000002 PSS35035.1 OIVN01001052 SPC89229.1 GEHC01000927 JAV46718.1 LBMM01007046 KMQ90088.1 ASHM01002084 PNY07765.1 JXUM01085595 KQ563513 KXJ73717.1 OIVN01003683 SPD12935.1 KQ971352 KYB26731.1 OIVN01002854 SPD06945.1 GEHC01000879 JAV46766.1 JYDO01000312 KRZ65659.1 OIVN01006239 SPD28653.1 GEZM01078128 GEZM01078127 JAV63002.1 PDCK01000039 PRQ60431.1 OIVN01000486 SPC81000.1 D83003 BAA11674.1 OIVN01001906 SPC98838.1 OIVN01001534 SPC95017.1 OIVN01005591 SPD23209.1 GFTR01008771 JAW07655.1 GBHO01024947 JAG18657.1 GBHO01024950 JAG18654.1 AM447500 CAN74198.1 JYDP01000350 KRZ01021.1 JYDN01000113 KRX56900.1 AM433482 CAN69340.1 OIVN01000036 SPC73022.1 OTG06758.1 JYDM01000185 KRZ84108.1 GBXI01007143 JAD07149.1
Proteomes
UP000053240
UP000218220
UP000036403
UP000055048
UP000054776
UP000054995
+ More
UP000054805 UP000054826 UP000054632 UP000075243 UP000084051 UP000215914 UP000002358 UP000054815 UP000054630 UP000241394 UP000236291 UP000069940 UP000249989 UP000007266 UP000054843 UP000238479 UP000055024 UP000054681 UP000054924
UP000054805 UP000054826 UP000054632 UP000075243 UP000084051 UP000215914 UP000002358 UP000054815 UP000054630 UP000241394 UP000236291 UP000069940 UP000249989 UP000007266 UP000054843 UP000238479 UP000055024 UP000054681 UP000054924
Pfam
Interpro
IPR025314
DUF4219
+ More
IPR012337 RNaseH-like_sf
IPR036875 Znf_CCHC_sf
IPR013103 RVT_2
IPR001584 Integrase_cat-core
IPR025724 GAG-pre-integrase_dom
IPR036397 RNaseH_sf
IPR001878 Znf_CCHC
IPR039537 Retrotran_Ty1/copia-like
IPR007123 Gelsolin-like_dom
IPR029006 ADF-H/Gelsolin-like_dom_sf
IPR036180 Gelsolin-like_dom_sf
IPR012990 Sec23_24_beta_S
IPR036291 NAD(P)-bd_dom_sf
IPR013740 Redoxin
IPR020904 Sc_DH/Rdtase_CS
IPR002347 SDR_fam
IPR036249 Thioredoxin-like_sf
IPR037944 PRX5-like
IPR025829 Zn_knuckle_CX2CX3GHX4C
IPR032675 LRR_dom_sf
IPR036576 WRKY_dom_sf
IPR001611 Leu-rich_rpt
IPR003657 WRKY_dom
IPR012337 RNaseH-like_sf
IPR036875 Znf_CCHC_sf
IPR013103 RVT_2
IPR001584 Integrase_cat-core
IPR025724 GAG-pre-integrase_dom
IPR036397 RNaseH_sf
IPR001878 Znf_CCHC
IPR039537 Retrotran_Ty1/copia-like
IPR007123 Gelsolin-like_dom
IPR029006 ADF-H/Gelsolin-like_dom_sf
IPR036180 Gelsolin-like_dom_sf
IPR012990 Sec23_24_beta_S
IPR036291 NAD(P)-bd_dom_sf
IPR013740 Redoxin
IPR020904 Sc_DH/Rdtase_CS
IPR002347 SDR_fam
IPR036249 Thioredoxin-like_sf
IPR037944 PRX5-like
IPR025829 Zn_knuckle_CX2CX3GHX4C
IPR032675 LRR_dom_sf
IPR036576 WRKY_dom_sf
IPR001611 Leu-rich_rpt
IPR003657 WRKY_dom
SUPFAM
Gene 3D
ProteinModelPortal
A0A194RU38
A0A2A4JRI1
A0A2A4J959
A0A194RJV9
A0A0J7NHJ8
A0A0A1WXQ3
+ More
A0A1Y1N932 A0A1W7R6K6 A0A1W7R6J6 A0A0N1ICY1 E5SB91 A0A0V0TUP0 A0A0V1APS7 A0A0A9WRE3 A0A1Y1MYL8 A0A1Y1MYL6 A0A224XA26 A0A1Y1LYU4 A0A0J7K725 A0A0A9WQD4 A0A1B6E9D0 B8YLY5 A0A0V1FKH2 A0A0V1K1Y9 A0A0V1F2E6 A0A151SDY6 B8YLY6 A0A151RJN4 B8YLY3 B8YLY4 A0A151R6U8 A0A151R2G6 A0A151TKT1 H6U1F0 A0A0P6IXK4 A0A2N9FMR2 A0A2N9GLP2 A0A2N9IUS6 Q6BCY1 P10978 H6U1E9 B8YLY7 A0A1Y1N664 A0A0A1XLZ7 A0A2S2NXV8 A0A251TN47 A0A1Y1N3G5 T1PKV2 A0A2M3YZL3 W8BWJ0 A0A034VP70 A0A2M3YZL7 A0A2M4CV35 A0A151RCT9 K7J8U2 B1N668 A0A0V1B5K0 A0A0V1B5A2 A0A2N9GHK9 A0A251TCA0 A0A0V0YNF4 A0A1Y1M571 B6V6Z8 A0A2N9ETC8 A0A151THM1 A0A2N9GY85 E5SJ15 A0A2N9JB15 A0A0V0RWJ3 A0A2R6RYD4 E5SZD6 A0A2N9FQJ1 A0A1W7R6C8 A0A0J7KI47 A0A2K3NXK0 A0A182GSW5 A0A2N9HLU0 A0A139WFV5 A0A2N9H4V1 A0A1W7R6H7 A0A0V1M1K0 A0A2N9IWX1 A0A1Y1KNM9 A0A2P6SP23 A0A2N9F1W0 Q9ZRJ0 A0A2N9GHB5 A0A2N9FWL6 A0A2N9IGN3 A0A224X581 A0A0A9XD75 A0A0A9XFC8 A5B5N4 A0A0V1GS48 A0A0V0V098 A5AS37 A0A2N9EE21 A0A251T6N3 A0A0V1NJN8 A0A0A1X768
A0A1Y1N932 A0A1W7R6K6 A0A1W7R6J6 A0A0N1ICY1 E5SB91 A0A0V0TUP0 A0A0V1APS7 A0A0A9WRE3 A0A1Y1MYL8 A0A1Y1MYL6 A0A224XA26 A0A1Y1LYU4 A0A0J7K725 A0A0A9WQD4 A0A1B6E9D0 B8YLY5 A0A0V1FKH2 A0A0V1K1Y9 A0A0V1F2E6 A0A151SDY6 B8YLY6 A0A151RJN4 B8YLY3 B8YLY4 A0A151R6U8 A0A151R2G6 A0A151TKT1 H6U1F0 A0A0P6IXK4 A0A2N9FMR2 A0A2N9GLP2 A0A2N9IUS6 Q6BCY1 P10978 H6U1E9 B8YLY7 A0A1Y1N664 A0A0A1XLZ7 A0A2S2NXV8 A0A251TN47 A0A1Y1N3G5 T1PKV2 A0A2M3YZL3 W8BWJ0 A0A034VP70 A0A2M3YZL7 A0A2M4CV35 A0A151RCT9 K7J8U2 B1N668 A0A0V1B5K0 A0A0V1B5A2 A0A2N9GHK9 A0A251TCA0 A0A0V0YNF4 A0A1Y1M571 B6V6Z8 A0A2N9ETC8 A0A151THM1 A0A2N9GY85 E5SJ15 A0A2N9JB15 A0A0V0RWJ3 A0A2R6RYD4 E5SZD6 A0A2N9FQJ1 A0A1W7R6C8 A0A0J7KI47 A0A2K3NXK0 A0A182GSW5 A0A2N9HLU0 A0A139WFV5 A0A2N9H4V1 A0A1W7R6H7 A0A0V1M1K0 A0A2N9IWX1 A0A1Y1KNM9 A0A2P6SP23 A0A2N9F1W0 Q9ZRJ0 A0A2N9GHB5 A0A2N9FWL6 A0A2N9IGN3 A0A224X581 A0A0A9XD75 A0A0A9XFC8 A5B5N4 A0A0V1GS48 A0A0V0V098 A5AS37 A0A2N9EE21 A0A251T6N3 A0A0V1NJN8 A0A0A1X768
PDB
5HRR
E-value=4.62195e-05,
Score=117
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Nucleus
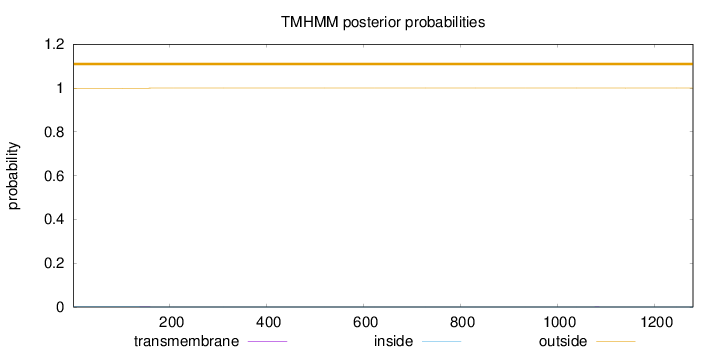
Length:
1280
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02335
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.00101
outside
1 - 1280
Population Genetic Test Statistics
Pi
0.05977
Theta
0.503898
Tajima's D
-2.147147
CLR
0.519727
CSRT
0
Interpretation
Uncertain
