Gene
KWMTBOMO00633
Pre Gene Modal
BGIBMGA003861
Annotation
PREDICTED:_nitrogen_permease_regulator_2-like_protein_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.872
Sequence
CDS
ATGGAAATCCCGACTTTACTATGTTCCTGGGAATGGCATCCAACTCTGTCAGACGTCCTGGAGATACTGTGTTCGCTGCAACAAGGTACAACCCTGAGGACTGTCAGCGACCGCTTCTCCAACTCTGCTCGACCCAACTTCGACATCAGACGACTGGTTGTCTTCGCTCAGATCCACGGCCTCATCAAATGCTTAAAAAGGTATCCAGTATATCTTCGAAATCCTCCTCGGCATAACGGTTTCAACACCCGCGTAGACCCCGTTCTCGGCATACGCAGACTGTTCACTGGCAAACATTGCGCCGATGAAATCTGCTGTCTCGCTCGCATCGACCTCCCGACCCTCGAGCAAATCATTGAAGAAGACCCGAACGTCGCTATTATATGGAGATAG
Protein
MEIPTLLCSWEWHPTLSDVLEILCSLQQGTTLRTVSDRFSNSARPNFDIRRLVVFAQIHGLIKCLKRYPVYLRNPPRHNGFNTRVDPVLGIRRLFTGKHCADEICCLARIDLPTLEQIIEEDPNVAIIWR
Summary
Uniprot
H9J2X4
A0A2H1V886
A0A2A4IVX4
A0A2A4IXB1
A0A2W1B3Q7
A0A212EQJ6
+ More
A0A194PG93 A0A067R7Y0 A0A336MBZ0 A0A0L7QK55 A0A1B6E1Q7 A7TA00 A0A1B6CWT5 A0A0A1XIV2 A0A2A3EM38 A0A2J7PXT5 A0A087ZVT4 A0A1A9YF26 A0A1A9VUK7 A0A1B0AMR6 A0A1B0A0V7 A0A0K8UFV4 A0A2I4B290 A0A034WGG6 A0A1I8MUY4 A0A1I8Q9U5 A0A1L1RSI4 A0A096M737 A0A2I4B293 A0A3B4CEV6 A0A0E9WG76 A0A3P9NBL6 A0A3B3R734 A0A1S3P243 A0A0S7LBH1 B4M2A5 A0A2D4F7T4 Q0P403 B4L1V0 A0A3B3XM59 A0A3P9NBM5 A0A3P9LG96 A0A3B3UZS8 A0A146NN69 A0A2U9BFP9 H2LFJ7 G3N8N0 A0A3P9I6X1 A0A1W4YXM5 A0A3P9MMZ9 A0A0M3QYV4 U3J5S2 A0A0P7YHT0 A0A1S3P247 A0A3B5M354 A0A3B3I132 A0A182S606 B4NCB0 A0A182N1A1 A0A310SKD5 F1NY69 A0A3L8R9X8 A0A218UQW2 A0A3M0JYL9 A0A1V4K8R5 A0A2U9BF21 A0A2I0TES9 A0A146P499 A0A2I0LHE3 A0A3P9JID9 M4AR75 W8BQH1 A0A3B4T4H4 A0A2U9BFS4 A0A182QPI2 A0A3P8RWK1 A7RTX3 A0A1W4YNM8 A0A3B4ZQV7 U3K2Y5 A0A3B3R757 B4Q1W7 A0A2B4RHX5 A0A3P8RVT0 B3N0C1 A0A1A8FLC4 A0A1A7WVU8 A0A1A8PQB9 A0A1A8IJK2 A0A3B5A4B0 B4IEU6 A0A1A8QNI8 A0A182MBW1 A0A182XXU5 A0A182W5G9 B4R647 A0A1A8CQH5
A0A194PG93 A0A067R7Y0 A0A336MBZ0 A0A0L7QK55 A0A1B6E1Q7 A7TA00 A0A1B6CWT5 A0A0A1XIV2 A0A2A3EM38 A0A2J7PXT5 A0A087ZVT4 A0A1A9YF26 A0A1A9VUK7 A0A1B0AMR6 A0A1B0A0V7 A0A0K8UFV4 A0A2I4B290 A0A034WGG6 A0A1I8MUY4 A0A1I8Q9U5 A0A1L1RSI4 A0A096M737 A0A2I4B293 A0A3B4CEV6 A0A0E9WG76 A0A3P9NBL6 A0A3B3R734 A0A1S3P243 A0A0S7LBH1 B4M2A5 A0A2D4F7T4 Q0P403 B4L1V0 A0A3B3XM59 A0A3P9NBM5 A0A3P9LG96 A0A3B3UZS8 A0A146NN69 A0A2U9BFP9 H2LFJ7 G3N8N0 A0A3P9I6X1 A0A1W4YXM5 A0A3P9MMZ9 A0A0M3QYV4 U3J5S2 A0A0P7YHT0 A0A1S3P247 A0A3B5M354 A0A3B3I132 A0A182S606 B4NCB0 A0A182N1A1 A0A310SKD5 F1NY69 A0A3L8R9X8 A0A218UQW2 A0A3M0JYL9 A0A1V4K8R5 A0A2U9BF21 A0A2I0TES9 A0A146P499 A0A2I0LHE3 A0A3P9JID9 M4AR75 W8BQH1 A0A3B4T4H4 A0A2U9BFS4 A0A182QPI2 A0A3P8RWK1 A7RTX3 A0A1W4YNM8 A0A3B4ZQV7 U3K2Y5 A0A3B3R757 B4Q1W7 A0A2B4RHX5 A0A3P8RVT0 B3N0C1 A0A1A8FLC4 A0A1A7WVU8 A0A1A8PQB9 A0A1A8IJK2 A0A3B5A4B0 B4IEU6 A0A1A8QNI8 A0A182MBW1 A0A182XXU5 A0A182W5G9 B4R647 A0A1A8CQH5
Pubmed
EMBL
BABH01031237
ODYU01001172
SOQ37006.1
NWSH01005590
PCG64117.1
PCG64116.1
+ More
KZ150373 PZC71129.1 AGBW02013268 OWR43773.1 KQ459605 KPI92048.1 KK852819 KDR15634.1 UFQT01000673 UFQT01000883 SSX26373.1 SSX27834.1 KQ415041 KOC58901.1 GEDC01005438 JAS31860.1 DS473722 EDO27172.1 GEDC01019358 JAS17940.1 GBXI01003415 JAD10877.1 KZ288219 PBC32229.1 NEVH01020852 PNF21147.1 JXJN01000553 JXJN01000554 JXJN01000555 GDHF01027109 JAI25205.1 GAKP01005748 GAKP01005747 JAC53205.1 AADN05000356 AYCK01000398 GBXM01019188 JAH89389.1 GBYX01177475 JAO86808.1 CH940651 EDW65809.1 IACJ01053808 LAA43565.1 BX004829 BX928750 BC122356 BC152088 AAI22357.1 CH933810 EDW06753.1 GCES01153440 JAQ32882.1 CP026248 AWP02639.1 CP012528 ALC48269.1 ADON01110048 JARO02005441 KPP66782.1 CH964239 EDW82469.1 KQ763450 OAD55051.1 QUSF01000740 RLV76298.1 MUZQ01000178 OWK56008.1 QRBI01000145 RMC00037.1 LSYS01004144 OPJ80846.1 AWP02637.1 AWP02638.1 KZ511511 PKU32297.1 GCES01147531 GCES01146564 GCES01064367 JAQ38791.1 JAR21956.1 AKCR02000708 PKK16857.1 GAMC01007492 JAB99063.1 AWP02640.1 AXCN02001237 DS469538 EDO45161.1 AGTO01011006 CM000162 EDX02542.1 LSMT01000601 PFX15865.1 CH902640 EDV38325.1 HAEB01013039 HAEC01003256 SBQ59566.1 HADW01008420 SBP09820.1 HAEG01009044 SBR83466.1 HAED01010972 HAEE01015466 SBQ97253.1 CH480832 EDW46200.1 HAEH01012555 HAEI01015755 SBR95082.1 AXCM01004309 CM000366 EDX18151.1 HADZ01018086 HAEA01003704 SBP82027.1
KZ150373 PZC71129.1 AGBW02013268 OWR43773.1 KQ459605 KPI92048.1 KK852819 KDR15634.1 UFQT01000673 UFQT01000883 SSX26373.1 SSX27834.1 KQ415041 KOC58901.1 GEDC01005438 JAS31860.1 DS473722 EDO27172.1 GEDC01019358 JAS17940.1 GBXI01003415 JAD10877.1 KZ288219 PBC32229.1 NEVH01020852 PNF21147.1 JXJN01000553 JXJN01000554 JXJN01000555 GDHF01027109 JAI25205.1 GAKP01005748 GAKP01005747 JAC53205.1 AADN05000356 AYCK01000398 GBXM01019188 JAH89389.1 GBYX01177475 JAO86808.1 CH940651 EDW65809.1 IACJ01053808 LAA43565.1 BX004829 BX928750 BC122356 BC152088 AAI22357.1 CH933810 EDW06753.1 GCES01153440 JAQ32882.1 CP026248 AWP02639.1 CP012528 ALC48269.1 ADON01110048 JARO02005441 KPP66782.1 CH964239 EDW82469.1 KQ763450 OAD55051.1 QUSF01000740 RLV76298.1 MUZQ01000178 OWK56008.1 QRBI01000145 RMC00037.1 LSYS01004144 OPJ80846.1 AWP02637.1 AWP02638.1 KZ511511 PKU32297.1 GCES01147531 GCES01146564 GCES01064367 JAQ38791.1 JAR21956.1 AKCR02000708 PKK16857.1 GAMC01007492 JAB99063.1 AWP02640.1 AXCN02001237 DS469538 EDO45161.1 AGTO01011006 CM000162 EDX02542.1 LSMT01000601 PFX15865.1 CH902640 EDV38325.1 HAEB01013039 HAEC01003256 SBQ59566.1 HADW01008420 SBP09820.1 HAEG01009044 SBR83466.1 HAED01010972 HAEE01015466 SBQ97253.1 CH480832 EDW46200.1 HAEH01012555 HAEI01015755 SBR95082.1 AXCM01004309 CM000366 EDX18151.1 HADZ01018086 HAEA01003704 SBP82027.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000027135
UP000053825
+ More
UP000001593 UP000242457 UP000235965 UP000005203 UP000092443 UP000078200 UP000092460 UP000092445 UP000192220 UP000095301 UP000095300 UP000000539 UP000028760 UP000261440 UP000242638 UP000261540 UP000087266 UP000008792 UP000000437 UP000009192 UP000261480 UP000265180 UP000261500 UP000246464 UP000001038 UP000007635 UP000265200 UP000192224 UP000092553 UP000016666 UP000034805 UP000261380 UP000075901 UP000007798 UP000075884 UP000276834 UP000197619 UP000269221 UP000190648 UP000053872 UP000002852 UP000261420 UP000075886 UP000265080 UP000261400 UP000016665 UP000002282 UP000225706 UP000007801 UP000001292 UP000075883 UP000076408 UP000075920 UP000000304
UP000001593 UP000242457 UP000235965 UP000005203 UP000092443 UP000078200 UP000092460 UP000092445 UP000192220 UP000095301 UP000095300 UP000000539 UP000028760 UP000261440 UP000242638 UP000261540 UP000087266 UP000008792 UP000000437 UP000009192 UP000261480 UP000265180 UP000261500 UP000246464 UP000001038 UP000007635 UP000265200 UP000192224 UP000092553 UP000016666 UP000034805 UP000261380 UP000075901 UP000007798 UP000075884 UP000276834 UP000197619 UP000269221 UP000190648 UP000053872 UP000002852 UP000261420 UP000075886 UP000265080 UP000261400 UP000016665 UP000002282 UP000225706 UP000007801 UP000001292 UP000075883 UP000076408 UP000075920 UP000000304
PRIDE
Pfam
PF06218 NPR2
Interpro
IPR009348
NPR2
ProteinModelPortal
H9J2X4
A0A2H1V886
A0A2A4IVX4
A0A2A4IXB1
A0A2W1B3Q7
A0A212EQJ6
+ More
A0A194PG93 A0A067R7Y0 A0A336MBZ0 A0A0L7QK55 A0A1B6E1Q7 A7TA00 A0A1B6CWT5 A0A0A1XIV2 A0A2A3EM38 A0A2J7PXT5 A0A087ZVT4 A0A1A9YF26 A0A1A9VUK7 A0A1B0AMR6 A0A1B0A0V7 A0A0K8UFV4 A0A2I4B290 A0A034WGG6 A0A1I8MUY4 A0A1I8Q9U5 A0A1L1RSI4 A0A096M737 A0A2I4B293 A0A3B4CEV6 A0A0E9WG76 A0A3P9NBL6 A0A3B3R734 A0A1S3P243 A0A0S7LBH1 B4M2A5 A0A2D4F7T4 Q0P403 B4L1V0 A0A3B3XM59 A0A3P9NBM5 A0A3P9LG96 A0A3B3UZS8 A0A146NN69 A0A2U9BFP9 H2LFJ7 G3N8N0 A0A3P9I6X1 A0A1W4YXM5 A0A3P9MMZ9 A0A0M3QYV4 U3J5S2 A0A0P7YHT0 A0A1S3P247 A0A3B5M354 A0A3B3I132 A0A182S606 B4NCB0 A0A182N1A1 A0A310SKD5 F1NY69 A0A3L8R9X8 A0A218UQW2 A0A3M0JYL9 A0A1V4K8R5 A0A2U9BF21 A0A2I0TES9 A0A146P499 A0A2I0LHE3 A0A3P9JID9 M4AR75 W8BQH1 A0A3B4T4H4 A0A2U9BFS4 A0A182QPI2 A0A3P8RWK1 A7RTX3 A0A1W4YNM8 A0A3B4ZQV7 U3K2Y5 A0A3B3R757 B4Q1W7 A0A2B4RHX5 A0A3P8RVT0 B3N0C1 A0A1A8FLC4 A0A1A7WVU8 A0A1A8PQB9 A0A1A8IJK2 A0A3B5A4B0 B4IEU6 A0A1A8QNI8 A0A182MBW1 A0A182XXU5 A0A182W5G9 B4R647 A0A1A8CQH5
A0A194PG93 A0A067R7Y0 A0A336MBZ0 A0A0L7QK55 A0A1B6E1Q7 A7TA00 A0A1B6CWT5 A0A0A1XIV2 A0A2A3EM38 A0A2J7PXT5 A0A087ZVT4 A0A1A9YF26 A0A1A9VUK7 A0A1B0AMR6 A0A1B0A0V7 A0A0K8UFV4 A0A2I4B290 A0A034WGG6 A0A1I8MUY4 A0A1I8Q9U5 A0A1L1RSI4 A0A096M737 A0A2I4B293 A0A3B4CEV6 A0A0E9WG76 A0A3P9NBL6 A0A3B3R734 A0A1S3P243 A0A0S7LBH1 B4M2A5 A0A2D4F7T4 Q0P403 B4L1V0 A0A3B3XM59 A0A3P9NBM5 A0A3P9LG96 A0A3B3UZS8 A0A146NN69 A0A2U9BFP9 H2LFJ7 G3N8N0 A0A3P9I6X1 A0A1W4YXM5 A0A3P9MMZ9 A0A0M3QYV4 U3J5S2 A0A0P7YHT0 A0A1S3P247 A0A3B5M354 A0A3B3I132 A0A182S606 B4NCB0 A0A182N1A1 A0A310SKD5 F1NY69 A0A3L8R9X8 A0A218UQW2 A0A3M0JYL9 A0A1V4K8R5 A0A2U9BF21 A0A2I0TES9 A0A146P499 A0A2I0LHE3 A0A3P9JID9 M4AR75 W8BQH1 A0A3B4T4H4 A0A2U9BFS4 A0A182QPI2 A0A3P8RWK1 A7RTX3 A0A1W4YNM8 A0A3B4ZQV7 U3K2Y5 A0A3B3R757 B4Q1W7 A0A2B4RHX5 A0A3P8RVT0 B3N0C1 A0A1A8FLC4 A0A1A7WVU8 A0A1A8PQB9 A0A1A8IJK2 A0A3B5A4B0 B4IEU6 A0A1A8QNI8 A0A182MBW1 A0A182XXU5 A0A182W5G9 B4R647 A0A1A8CQH5
PDB
6CET
E-value=4.78218e-12,
Score=165
Ontologies
KEGG
GO
PANTHER
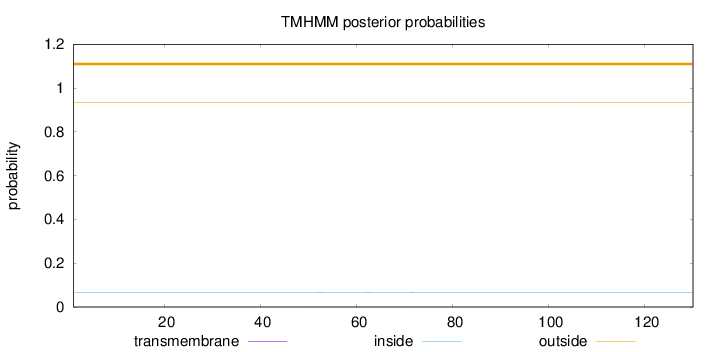
Topology
Length:
130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00866
Exp number, first 60 AAs:
0.00487
Total prob of N-in:
0.06575
outside
1 - 130
Population Genetic Test Statistics
Pi
5.840346
Theta
5.167931
Tajima's D
0.11455
CLR
0.005575
CSRT
0.424878756062197
Interpretation
Possibly Positive selection
