Gene
KWMTBOMO00620
Pre Gene Modal
BGIBMGA003854
Annotation
PREDICTED:_proton-coupled_amino_acid_transporter_4_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.955
Sequence
CDS
ATGATCAACCCTTCCTTCGAACCGGACGATTTTGTTCCACACAGCCTATCACCGGTGAAGGATGAAAAGAAGAAGCCTGGCGATAAAAGCATCTTCTTGGTCAACATGAAGGAGAAGGACCTTCAAGAAGTAGAGGAATACGAACCGTATGACAACAGGGTCGTTGACCATCCCACAACGAACACGGAGACCCTACTTCATCTTCTGAAAGGCAGTTTAGGCACTGGTATTCTGGCAATGCCGCGTGCCTTCTCCCATGCCGGATATGTTGTAGGCGGCATAGGCACTGTGATTATCGGTGTGCTGTGTACTTACTGCATCCACATATTACTGGATTCTTGCTATGCGCTGTGCAAGAGGCGGAAGGTGCCCTCCTTGACGTACACCGCTGCAGCAGAAGCGGCGCTCTCCGAGGGCCCGGACTGGTGCAAGGCATGCGCTCCTTACGCTGCGCTCATCGTGAACGTATTTCTGCTGATCTACCAGATAGGCACGTGTTGCGTGTACGTTGTTTTCGTTTCTGAGAACATCCATTTCGTCTTGGTGAAGCGGTTCAACGTGGAAATCACCGTCTTCCAAGTGATGTTGTGTATCCTGCTACCATTGATACTGATCAATTGGGTACGAGACCTGAAATATCTCGCTCCGTTCTCTGCTGTAGCCAATGTGGTGACTATAGTTGGATTCGGTATCATCTTGTACTACATATTCAGAGAAACACCGACTTTAGAAGGCAAAGAACCTGTTGGTCGGCTCGCTGACTTCCCCCTGTTCTTCGGCACCGTTCTATTCGCTCTGGAAGCCATTGGTGTGATTCTACCGTTGGAGAACGAAATGAAAACACCGAAGTCATTCGTCGGCAAGTTCGGAGTCTTGAATCGAGCCATGATTAGTATCATCATACTGTACGTCGGTATGGGATTGTTTGGATATCTGCGTTACGGGGAGGAGTCCATGGGCAGCATCACTTTGAACCTACCATCTGAAACAGAAACTCTTGCAAGTGTCGTACAGTGTCTCCTTGCATTCGCAATCTTCATAACTCACGGACTGGCTTGCTACGTCGCCATTGACATCCTCTGGAACGAATACATCGGTGTGCGACTACTGAACAGCAAGTTCCGTGTCGTATGGGAATATATTGTGAGGACTATCATCGTACTGATCACTTTCGGCTTAGCTGCGGCGGTTCCGGAATTGGATCTGTTCATTTCCCTGTTCGGTGCTCTGTGTCTGTCTGCTCTCGGGCTAGCATTCCCTGCCTTCATACAAAGCTGCACGTACTGGTACTATGTTTCACGCTCTGAACGCATTCGTATGATCATCAAGAACATCATTGTAGTGTTGTTCGGGGTTTTGGGCCTAATCGTCGGCACATGGACGTCACTGCAGAGCATCATCCAGAAGTTCTCAGGACCACACAATCCAATTGTCAACGGCACGCACTTTACAATGTTAAATGAAACAACGCTTCCTTGA
Protein
MINPSFEPDDFVPHSLSPVKDEKKKPGDKSIFLVNMKEKDLQEVEEYEPYDNRVVDHPTTNTETLLHLLKGSLGTGILAMPRAFSHAGYVVGGIGTVIIGVLCTYCIHILLDSCYALCKRRKVPSLTYTAAAEAALSEGPDWCKACAPYAALIVNVFLLIYQIGTCCVYVVFVSENIHFVLVKRFNVEITVFQVMLCILLPLILINWVRDLKYLAPFSAVANVVTIVGFGIILYYIFRETPTLEGKEPVGRLADFPLFFGTVLFALEAIGVILPLENEMKTPKSFVGKFGVLNRAMISIIILYVGMGLFGYLRYGEESMGSITLNLPSETETLASVVQCLLAFAIFITHGLACYVAIDILWNEYIGVRLLNSKFRVVWEYIVRTIIVLITFGLAAAVPELDLFISLFGALCLSALGLAFPAFIQSCTYWYYVSRSERIRMIIKNIIVVLFGVLGLIVGTWTSLQSIIQKFSGPHNPIVNGTHFTMLNETTLP
Summary
Uniprot
A0A0L7LNZ0
A0A194QMW5
A0A194PG38
A0A2H1V3G6
A0A2A4JZH8
A0A212F9N7
+ More
A0A1L8DEQ7 A0A1B0GIE1 A0A0Q9WWM3 B4LEG6 A0A0Q9WL13 A0A1Y0AWM7 B4N441 A0A182NAR6 A0A182TG07 A0A182UWB9 A0A182KTL7 A0A182HRI9 B0WVR1 A0A0M4EAI7 A0A1Q3FR37 A0A0A1WIX6 B4KWF6 A0A182W914 A0A182Q9U4 Q7Q198 A0A3B0KI52 A0A182RR73 A0A1J1IZ53 Q173C2 A0A3B0JTL1 A0A0L0C113 A0A182PCA9 A0A067RLL7 W8B327 Q16XD9 A0A1I8PLA8 A0A158NQR8 Q2LZY9 B4H935 A0A084W8L0 A0A034VCJ9 A0A195DI13 A0A182IUZ7 A0A2M3ZF73 A0A0R3P851 A0A182XJL2 A0A2M3YXZ0 A0A0K8WLN1 A0A1A9W4P7 A0A2M4BLY7 A0A182KAV7 A0A2J7PW22 A0A0L7R8S2 A0A151XAU2 W5JMH1 A0A182F3V0 A0A023EUH1 A0A182H343 A0A182G8X1 A0A2M4A082 A0A1A9YML1 B4J1Q4 A0A1W4W3Q7 A0A2A3E5F9 A0A0J7NX94 A0A1B0G5A8 A0A1A9ZWL7 A0A3B0JTH5 B3NCQ6 B4HDU3 B4QPM8 Q9VTD7 Q7YTZ0 D3DME7 B3M5N3 D6WF64 A0A0P8XVE0 A0A088AIY5 B4PEJ7 A0A182MCC9 A0A182YR04 T1E1X8 A0A1I8MBP9 E2BM88 T1PCX6 A0A1Y1LA59 A0A026VX47 V5I8Q8 A0A1W4WZ09 A0A0K8VJA6 E0VZX4 F4WKB6 A0A0T6BGT5 A0A1B0D728 A0A2P8ZEK8 K7IR10 A0A195EQQ6
A0A1L8DEQ7 A0A1B0GIE1 A0A0Q9WWM3 B4LEG6 A0A0Q9WL13 A0A1Y0AWM7 B4N441 A0A182NAR6 A0A182TG07 A0A182UWB9 A0A182KTL7 A0A182HRI9 B0WVR1 A0A0M4EAI7 A0A1Q3FR37 A0A0A1WIX6 B4KWF6 A0A182W914 A0A182Q9U4 Q7Q198 A0A3B0KI52 A0A182RR73 A0A1J1IZ53 Q173C2 A0A3B0JTL1 A0A0L0C113 A0A182PCA9 A0A067RLL7 W8B327 Q16XD9 A0A1I8PLA8 A0A158NQR8 Q2LZY9 B4H935 A0A084W8L0 A0A034VCJ9 A0A195DI13 A0A182IUZ7 A0A2M3ZF73 A0A0R3P851 A0A182XJL2 A0A2M3YXZ0 A0A0K8WLN1 A0A1A9W4P7 A0A2M4BLY7 A0A182KAV7 A0A2J7PW22 A0A0L7R8S2 A0A151XAU2 W5JMH1 A0A182F3V0 A0A023EUH1 A0A182H343 A0A182G8X1 A0A2M4A082 A0A1A9YML1 B4J1Q4 A0A1W4W3Q7 A0A2A3E5F9 A0A0J7NX94 A0A1B0G5A8 A0A1A9ZWL7 A0A3B0JTH5 B3NCQ6 B4HDU3 B4QPM8 Q9VTD7 Q7YTZ0 D3DME7 B3M5N3 D6WF64 A0A0P8XVE0 A0A088AIY5 B4PEJ7 A0A182MCC9 A0A182YR04 T1E1X8 A0A1I8MBP9 E2BM88 T1PCX6 A0A1Y1LA59 A0A026VX47 V5I8Q8 A0A1W4WZ09 A0A0K8VJA6 E0VZX4 F4WKB6 A0A0T6BGT5 A0A1B0D728 A0A2P8ZEK8 K7IR10 A0A195EQQ6
Pubmed
26227816
26354079
22118469
17994087
18057021
28341416
+ More
20966253 25830018 12364791 17510324 19801431 26108605 24845553 24495485 21347285 15632085 24438588 25348373 20920257 23761445 24945155 26483478 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 17550304 25244985 24330624 25315136 20798317 28004739 24508170 30249741 20566863 21719571 29403074 20075255
20966253 25830018 12364791 17510324 19801431 26108605 24845553 24495485 21347285 15632085 24438588 25348373 20920257 23761445 24945155 26483478 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 17550304 25244985 24330624 25315136 20798317 28004739 24508170 30249741 20566863 21719571 29403074 20075255
EMBL
JTDY01000490
KOB76941.1
KQ461196
KPJ06310.1
KQ459605
KPI91993.1
+ More
ODYU01000310 SOQ34824.1 NWSH01000355 PCG77088.1 AGBW02009564 OWR50447.1 GFDF01009158 JAV04926.1 AJWK01013779 CH940647 KRF84725.1 EDW70142.1 KRF84723.1 KRF84724.1 KRF84726.1 KY921804 ART29393.1 CH964095 EDW78915.1 APCN01001708 DS232131 EDS35668.1 CP012525 ALC44278.1 GFDL01004996 JAV30049.1 GBXI01015812 JAC98479.1 CH933809 EDW19585.1 KRG06698.1 AXCN02000660 AAAB01008980 EAA14378.3 OUUW01000009 SPP85436.1 CVRI01000063 CRL04446.1 GQ231545 CH477427 ACS96436.1 EAT41138.1 SPP85435.1 JRES01001060 KNC25941.1 KK852550 KDR21505.1 GAMC01011041 GAMC01011040 GAMC01011039 JAB95515.1 CH477541 EAT39273.1 ADTU01023342 ADTU01023343 ADTU01023344 CH379069 EAL31148.1 CH479226 EDW35250.1 ATLV01021489 KE525319 KFB46554.1 GAKP01018758 GAKP01018757 JAC40194.1 KQ980824 KYN12477.1 GGFM01006456 MBW27207.1 KRT07923.1 GGFM01000382 MBW21133.1 GDHF01006150 GDHF01000317 JAI46164.1 JAI51997.1 GGFJ01004946 MBW54087.1 NEVH01020938 PNF20533.1 KQ414632 KOC67166.1 KQ982335 KYQ57502.1 ADMH02000680 ETN65336.1 GAPW01001017 JAC12581.1 JXUM01024157 KQ560681 KXJ81293.1 JXUM01048337 JXUM01048338 KQ561558 KXJ78172.1 GGFK01000905 MBW34226.1 CH916366 EDV96974.1 KZ288372 PBC26724.1 LBMM01001038 KMQ97035.1 CCAG010002099 CCAG010002100 SPP85437.1 CH954178 EDV51353.2 KQS43784.1 CH480815 EDW41033.1 CM000363 CM002912 EDX10019.1 KMY98908.1 AE014296 AAF50114.1 BT010105 AAF50115.1 AAQ22574.1 BT100057 ACX61598.1 CH902618 EDV40667.1 KQ971327 EEZ99835.1 KPU78685.1 CM000159 EDW94063.2 KRK01639.1 AXCM01000563 GALA01001680 JAA93172.1 GL449158 EFN83214.1 KA646514 AFP61143.1 GEZM01063216 GEZM01063215 JAV69190.1 KK107648 QOIP01000008 EZA48332.1 RLU19972.1 GALX01004260 JAB64206.1 GDHF01013338 JAI38976.1 DS235854 EEB18929.1 GL888199 EGI65340.1 LJIG01000375 KRT86565.1 AJVK01012298 AJVK01012299 PYGN01000080 PSN54890.1 KQ982021 KYN30543.1
ODYU01000310 SOQ34824.1 NWSH01000355 PCG77088.1 AGBW02009564 OWR50447.1 GFDF01009158 JAV04926.1 AJWK01013779 CH940647 KRF84725.1 EDW70142.1 KRF84723.1 KRF84724.1 KRF84726.1 KY921804 ART29393.1 CH964095 EDW78915.1 APCN01001708 DS232131 EDS35668.1 CP012525 ALC44278.1 GFDL01004996 JAV30049.1 GBXI01015812 JAC98479.1 CH933809 EDW19585.1 KRG06698.1 AXCN02000660 AAAB01008980 EAA14378.3 OUUW01000009 SPP85436.1 CVRI01000063 CRL04446.1 GQ231545 CH477427 ACS96436.1 EAT41138.1 SPP85435.1 JRES01001060 KNC25941.1 KK852550 KDR21505.1 GAMC01011041 GAMC01011040 GAMC01011039 JAB95515.1 CH477541 EAT39273.1 ADTU01023342 ADTU01023343 ADTU01023344 CH379069 EAL31148.1 CH479226 EDW35250.1 ATLV01021489 KE525319 KFB46554.1 GAKP01018758 GAKP01018757 JAC40194.1 KQ980824 KYN12477.1 GGFM01006456 MBW27207.1 KRT07923.1 GGFM01000382 MBW21133.1 GDHF01006150 GDHF01000317 JAI46164.1 JAI51997.1 GGFJ01004946 MBW54087.1 NEVH01020938 PNF20533.1 KQ414632 KOC67166.1 KQ982335 KYQ57502.1 ADMH02000680 ETN65336.1 GAPW01001017 JAC12581.1 JXUM01024157 KQ560681 KXJ81293.1 JXUM01048337 JXUM01048338 KQ561558 KXJ78172.1 GGFK01000905 MBW34226.1 CH916366 EDV96974.1 KZ288372 PBC26724.1 LBMM01001038 KMQ97035.1 CCAG010002099 CCAG010002100 SPP85437.1 CH954178 EDV51353.2 KQS43784.1 CH480815 EDW41033.1 CM000363 CM002912 EDX10019.1 KMY98908.1 AE014296 AAF50114.1 BT010105 AAF50115.1 AAQ22574.1 BT100057 ACX61598.1 CH902618 EDV40667.1 KQ971327 EEZ99835.1 KPU78685.1 CM000159 EDW94063.2 KRK01639.1 AXCM01000563 GALA01001680 JAA93172.1 GL449158 EFN83214.1 KA646514 AFP61143.1 GEZM01063216 GEZM01063215 JAV69190.1 KK107648 QOIP01000008 EZA48332.1 RLU19972.1 GALX01004260 JAB64206.1 GDHF01013338 JAI38976.1 DS235854 EEB18929.1 GL888199 EGI65340.1 LJIG01000375 KRT86565.1 AJVK01012298 AJVK01012299 PYGN01000080 PSN54890.1 KQ982021 KYN30543.1
Proteomes
UP000037510
UP000053240
UP000053268
UP000218220
UP000007151
UP000092461
+ More
UP000008792 UP000007798 UP000075884 UP000075902 UP000075903 UP000075882 UP000075840 UP000002320 UP000092553 UP000009192 UP000075920 UP000075886 UP000007062 UP000268350 UP000075900 UP000183832 UP000008820 UP000037069 UP000075885 UP000027135 UP000095300 UP000005205 UP000001819 UP000008744 UP000030765 UP000078492 UP000075880 UP000076407 UP000091820 UP000075881 UP000235965 UP000053825 UP000075809 UP000000673 UP000069272 UP000069940 UP000249989 UP000092443 UP000001070 UP000192221 UP000242457 UP000036403 UP000092444 UP000092445 UP000008711 UP000001292 UP000000304 UP000000803 UP000007801 UP000007266 UP000005203 UP000002282 UP000075883 UP000076408 UP000095301 UP000008237 UP000053097 UP000279307 UP000192223 UP000009046 UP000007755 UP000092462 UP000245037 UP000002358 UP000078541
UP000008792 UP000007798 UP000075884 UP000075902 UP000075903 UP000075882 UP000075840 UP000002320 UP000092553 UP000009192 UP000075920 UP000075886 UP000007062 UP000268350 UP000075900 UP000183832 UP000008820 UP000037069 UP000075885 UP000027135 UP000095300 UP000005205 UP000001819 UP000008744 UP000030765 UP000078492 UP000075880 UP000076407 UP000091820 UP000075881 UP000235965 UP000053825 UP000075809 UP000000673 UP000069272 UP000069940 UP000249989 UP000092443 UP000001070 UP000192221 UP000242457 UP000036403 UP000092444 UP000092445 UP000008711 UP000001292 UP000000304 UP000000803 UP000007801 UP000007266 UP000005203 UP000002282 UP000075883 UP000076408 UP000095301 UP000008237 UP000053097 UP000279307 UP000192223 UP000009046 UP000007755 UP000092462 UP000245037 UP000002358 UP000078541
Pfam
PF01490 Aa_trans
Interpro
IPR013057
AA_transpt_TM
ProteinModelPortal
A0A0L7LNZ0
A0A194QMW5
A0A194PG38
A0A2H1V3G6
A0A2A4JZH8
A0A212F9N7
+ More
A0A1L8DEQ7 A0A1B0GIE1 A0A0Q9WWM3 B4LEG6 A0A0Q9WL13 A0A1Y0AWM7 B4N441 A0A182NAR6 A0A182TG07 A0A182UWB9 A0A182KTL7 A0A182HRI9 B0WVR1 A0A0M4EAI7 A0A1Q3FR37 A0A0A1WIX6 B4KWF6 A0A182W914 A0A182Q9U4 Q7Q198 A0A3B0KI52 A0A182RR73 A0A1J1IZ53 Q173C2 A0A3B0JTL1 A0A0L0C113 A0A182PCA9 A0A067RLL7 W8B327 Q16XD9 A0A1I8PLA8 A0A158NQR8 Q2LZY9 B4H935 A0A084W8L0 A0A034VCJ9 A0A195DI13 A0A182IUZ7 A0A2M3ZF73 A0A0R3P851 A0A182XJL2 A0A2M3YXZ0 A0A0K8WLN1 A0A1A9W4P7 A0A2M4BLY7 A0A182KAV7 A0A2J7PW22 A0A0L7R8S2 A0A151XAU2 W5JMH1 A0A182F3V0 A0A023EUH1 A0A182H343 A0A182G8X1 A0A2M4A082 A0A1A9YML1 B4J1Q4 A0A1W4W3Q7 A0A2A3E5F9 A0A0J7NX94 A0A1B0G5A8 A0A1A9ZWL7 A0A3B0JTH5 B3NCQ6 B4HDU3 B4QPM8 Q9VTD7 Q7YTZ0 D3DME7 B3M5N3 D6WF64 A0A0P8XVE0 A0A088AIY5 B4PEJ7 A0A182MCC9 A0A182YR04 T1E1X8 A0A1I8MBP9 E2BM88 T1PCX6 A0A1Y1LA59 A0A026VX47 V5I8Q8 A0A1W4WZ09 A0A0K8VJA6 E0VZX4 F4WKB6 A0A0T6BGT5 A0A1B0D728 A0A2P8ZEK8 K7IR10 A0A195EQQ6
A0A1L8DEQ7 A0A1B0GIE1 A0A0Q9WWM3 B4LEG6 A0A0Q9WL13 A0A1Y0AWM7 B4N441 A0A182NAR6 A0A182TG07 A0A182UWB9 A0A182KTL7 A0A182HRI9 B0WVR1 A0A0M4EAI7 A0A1Q3FR37 A0A0A1WIX6 B4KWF6 A0A182W914 A0A182Q9U4 Q7Q198 A0A3B0KI52 A0A182RR73 A0A1J1IZ53 Q173C2 A0A3B0JTL1 A0A0L0C113 A0A182PCA9 A0A067RLL7 W8B327 Q16XD9 A0A1I8PLA8 A0A158NQR8 Q2LZY9 B4H935 A0A084W8L0 A0A034VCJ9 A0A195DI13 A0A182IUZ7 A0A2M3ZF73 A0A0R3P851 A0A182XJL2 A0A2M3YXZ0 A0A0K8WLN1 A0A1A9W4P7 A0A2M4BLY7 A0A182KAV7 A0A2J7PW22 A0A0L7R8S2 A0A151XAU2 W5JMH1 A0A182F3V0 A0A023EUH1 A0A182H343 A0A182G8X1 A0A2M4A082 A0A1A9YML1 B4J1Q4 A0A1W4W3Q7 A0A2A3E5F9 A0A0J7NX94 A0A1B0G5A8 A0A1A9ZWL7 A0A3B0JTH5 B3NCQ6 B4HDU3 B4QPM8 Q9VTD7 Q7YTZ0 D3DME7 B3M5N3 D6WF64 A0A0P8XVE0 A0A088AIY5 B4PEJ7 A0A182MCC9 A0A182YR04 T1E1X8 A0A1I8MBP9 E2BM88 T1PCX6 A0A1Y1LA59 A0A026VX47 V5I8Q8 A0A1W4WZ09 A0A0K8VJA6 E0VZX4 F4WKB6 A0A0T6BGT5 A0A1B0D728 A0A2P8ZEK8 K7IR10 A0A195EQQ6
Ontologies
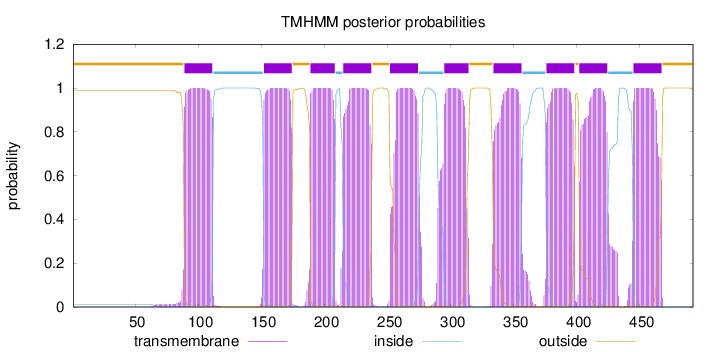
Topology
Length:
492
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
223.00192
Exp number, first 60 AAs:
0.00025
Total prob of N-in:
0.01276
outside
1 - 88
TMhelix
89 - 111
inside
112 - 151
TMhelix
152 - 174
outside
175 - 188
TMhelix
189 - 208
inside
209 - 214
TMhelix
215 - 237
outside
238 - 251
TMhelix
252 - 274
inside
275 - 294
TMhelix
295 - 314
outside
315 - 333
TMhelix
334 - 356
inside
357 - 375
TMhelix
376 - 398
outside
399 - 401
TMhelix
402 - 424
inside
425 - 444
TMhelix
445 - 467
outside
468 - 492
Population Genetic Test Statistics
Pi
24.503211
Theta
20.960169
Tajima's D
-1.97413
CLR
0.026067
CSRT
0.0170491475426229
Interpretation
Uncertain
