Gene
KWMTBOMO00618
Pre Gene Modal
BGIBMGA003852
Annotation
PREDICTED:_uncharacterized_protein_LOC101746464_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.702
Sequence
CDS
ATGTCAGAACCCATCATTGTCACCATAGATACCATAGAATGCGTTCTAAGTCTCAATCCTCCTGCACCAAATGAAGAGACCGTACCTCCTGACTCTCCAAGCAGACGAGCGCAAGTCGTAGAAGCACCTCCGGGTTACATGCAAGCATTAGTGCGCCGTATCGTGAGCAACATAAGCCTCCGAATACATCACCTCATCGTGAAATATGTGCAAGATGATATCGTAATGTCACTGAATGTGAAACATCTAGCTGTCGACAGTGCCGACTCGGATTGGGAACCGTCTTTTGCTGATATCGATCAGAACCTGCCCATCATCAGGAGGCTCGTACATCTAGATGACCTAACCCTCTGCCTGGACAGAGCTGATTCTGATGGCAAGATAAGATTCTACCAGGAACCTTTGCTGTATCGATGCCAATTAGAGTTAAGAGTACTGACCCGTTTAGTATCAGCGAATACACGTCGCGCAAGCAGCTTGAGTGTCCAGCTTCGATCAAGTAAACTGGCTTGGGGCGTGACCAGTGACCAACTGACTTTACTCTTGCGATTGATCAGAGAACGATCTGTGGTCAAGGTGCAACCGCAAACCACCAAGATTATCAATATCAACCAACCGCCAGCGCATTTAACGAGCTCAAGTTCATCTGAGCCGGCTCGTACCGAGAGCTGGTCGGAGTGGGCGTGGTCTTGGTTCCCTATGACATCTGATGCTGAGGGTGGAATAGAAGAAGAACGCATGCCTCCGGCACCTACACCTCTTCATTTTAACGCCTATTTCGATGACATATCCCTTGTTTTCAAGGTAATGGAATCGGAGTCAAGTACTCGTAAACGTTCACGTGGCGTGTTAGAGCTGTGCGCTTCACACGGCGCCATCAAGAGCTACGTTTGCAGACCGACTTCGTTCCGGATGCAAATGGCGACGCGCACGTTAACGATCAAATCGTCCGGAAGATGTGTCTGCGGACATCTCGATTACACTATGGGTAAAGACGAGCCTACCGTGTACCTGTGTAAATCGGAGAGCGAGCGCGTCGACAACTGGACCTGGCCCGCGAATGAATTCAGCGGAGGTCACGTGGAGACAGCGGAAGCAGCTCAGGAAACTCCTAAGGACGAGAACCAAAATCCGGAACCGATTATAGACGAGTCGGATCAATCAGGACACTCCACGCCGGAGCCCAAAAATAACATCGACGAAGATGACAAACTGTGGGCACAGATGTCGCCACTGTTGTACGTTAATTACACTCATGAACGATCACCGCCCGACGTTTACGAAAACCCTTATGAACATCCGCCTGAAGATTTTCAATATAGTGATTGGGCAGAAGAAAGCAACCTGAAAATCGAAATTGAATCTTTAGATATCAAACTCAGTACCGGATTGCTGCATAGACTTTCAGCCTTGAAGAACATTTACAAAGAGCTACCGCCAGTTCCCGACGTTGAACTACCAATGCGCGTGCTGACAGTCGAAGAATGCGACGCTCTATTCGACAATCTACCCTTGCGGCGTTTGTCTGTTGAAATGAAAGGGCTCAACGTCCGCGTACACCCTTTCGATCACTCGGCACAGGACCGTACTCCGTCGCACCCGGTCGTTTTGGAAGTCTACATGCCGAGGGGCATCCTGATTGTCAGTGGACCTCTGTATCCTCATAGAGTCTGTTCGGCGGCTTGTCAGATGCCGGATGACGCGGGACCGCTATGGCAAGGGGCACGTGTTCACGTGTCCGCCATTTTGTCCGGTATTGAGGCTAGCATTTGCTCCGTCGACGGGCAGACTCGGCCGTGCGCGAGAACCGATCTCCGCATAGTTGCGCACTCGCTTCAGTACAGAGAATTCTTCACGAAGCGGGAGAGCGTTGTCTTCAGTTATTCAATTAAGATGCGTAAGTAA
Protein
MSEPIIVTIDTIECVLSLNPPAPNEETVPPDSPSRRAQVVEAPPGYMQALVRRIVSNISLRIHHLIVKYVQDDIVMSLNVKHLAVDSADSDWEPSFADIDQNLPIIRRLVHLDDLTLCLDRADSDGKIRFYQEPLLYRCQLELRVLTRLVSANTRRASSLSVQLRSSKLAWGVTSDQLTLLLRLIRERSVVKVQPQTTKIININQPPAHLTSSSSSEPARTESWSEWAWSWFPMTSDAEGGIEEERMPPAPTPLHFNAYFDDISLVFKVMESESSTRKRSRGVLELCASHGAIKSYVCRPTSFRMQMATRTLTIKSSGRCVCGHLDYTMGKDEPTVYLCKSESERVDNWTWPANEFSGGHVETAEAAQETPKDENQNPEPIIDESDQSGHSTPEPKNNIDEDDKLWAQMSPLLYVNYTHERSPPDVYENPYEHPPEDFQYSDWAEESNLKIEIESLDIKLSTGLLHRLSALKNIYKELPPVPDVELPMRVLTVEECDALFDNLPLRRLSVEMKGLNVRVHPFDHSAQDRTPSHPVVLEVYMPRGILIVSGPLYPHRVCSAACQMPDDAGPLWQGARVHVSAILSGIEASICSVDGQTRPCARTDLRIVAHSLQYREFFTKRESVVFSYSIKMRK
Summary
Uniprot
H9J2W5
A0A212EGN6
A0A2A4JQH4
A0A2A4JQ03
A0A194PLL2
A0A194QL96
+ More
A0A158NQI3 A0A182J7D6 A0A0M9ABX8 A0A1Y1JZC7 A0A182Q1H4 A0A182P8W4 A0A182KX82 A0A182HJY7 A0A182FKZ1 A0A182WW13 Q7QH31 A0A151IUV9 A0A195FGP8 A0A182MEU4 A0A182NL10 A0A182UW75 A0A182JVY1 A0A151WEG8 A0A154P9M1 A0A0C9QMT7 A0A182U8R3 A0A182R877 A0A182VS60 A0A0J7KZS0 A0A151IDA6 A0A088ADA9 A0A0L7RJC9 A0A182YLN0 B0WZW5 U4UBA7 N6UX27 A0A067RGP1 A0A3L8DZI2 E2BJX3 A0A026W8L7 Q16SU8 N6UWR0 D6X2R8 W5J3V5 A0A195BDS5 A0A139WBB9 A0A232EXR6 A0A1J1J4M2 F4WJK5 A0A131YX57 K7JAY2 A0A310SR06 A0A2S2P3S9 A0A2A3EFC8 A0A0K2T0P4 J9JMF7 A0A3M6U0T9 A0A0P5K9L6 A0A0P6GL47 A0A0P6GPY1 A0A0P5LR64 A0A0P6FAD3 A0A0P5IS95 A0A0P5IN93 A0A0P5SEJ8 A0A0N8C843 A0A0P6HP95 A0A0P5PUH7 A0A0P6C7M5 A0A0P5PJ39 A0A0N8DGM7 A0A164NXP5 A0A2R5L9N5
A0A158NQI3 A0A182J7D6 A0A0M9ABX8 A0A1Y1JZC7 A0A182Q1H4 A0A182P8W4 A0A182KX82 A0A182HJY7 A0A182FKZ1 A0A182WW13 Q7QH31 A0A151IUV9 A0A195FGP8 A0A182MEU4 A0A182NL10 A0A182UW75 A0A182JVY1 A0A151WEG8 A0A154P9M1 A0A0C9QMT7 A0A182U8R3 A0A182R877 A0A182VS60 A0A0J7KZS0 A0A151IDA6 A0A088ADA9 A0A0L7RJC9 A0A182YLN0 B0WZW5 U4UBA7 N6UX27 A0A067RGP1 A0A3L8DZI2 E2BJX3 A0A026W8L7 Q16SU8 N6UWR0 D6X2R8 W5J3V5 A0A195BDS5 A0A139WBB9 A0A232EXR6 A0A1J1J4M2 F4WJK5 A0A131YX57 K7JAY2 A0A310SR06 A0A2S2P3S9 A0A2A3EFC8 A0A0K2T0P4 J9JMF7 A0A3M6U0T9 A0A0P5K9L6 A0A0P6GL47 A0A0P6GPY1 A0A0P5LR64 A0A0P6FAD3 A0A0P5IS95 A0A0P5IN93 A0A0P5SEJ8 A0A0N8C843 A0A0P6HP95 A0A0P5PUH7 A0A0P6C7M5 A0A0P5PJ39 A0A0N8DGM7 A0A164NXP5 A0A2R5L9N5
Pubmed
EMBL
BABH01031270
BABH01031271
BABH01031272
BABH01031273
BABH01031274
BABH01031275
+ More
BABH01031276 BABH01031277 AGBW02015068 OWR40641.1 NWSH01000888 PCG73672.1 PCG73674.1 KQ459605 KPI91995.1 KQ461196 KPJ06307.1 ADTU01000400 KQ435706 KOX80133.1 GEZM01099854 JAV53140.1 AXCN02000090 APCN01002223 AAAB01008820 EAA05407.5 KQ980937 KYN11280.1 KQ981606 KYN39551.1 AXCM01000231 AXCM01000232 AXCM01000233 KQ983238 KYQ46225.1 KQ434851 KZC08609.1 GBYB01004819 JAG74586.1 LBMM01001707 KMQ95861.1 KQ977978 KYM98381.1 KQ414581 KOC70975.1 DS232219 EDS37790.1 KB631924 ERL87250.1 APGK01002955 KB735045 ENN83457.1 KK852480 KDR22947.1 QOIP01000002 RLU25817.1 GL448708 EFN83982.1 KK107391 EZA51369.1 CH477665 EAT37541.1 APGK01004030 KB735543 ENN83352.1 KQ971372 EFA10287.2 ADMH02002130 ETN58526.1 KQ976509 KYM82708.1 KYB25206.1 NNAY01001682 OXU23234.1 CVRI01000065 CRL05825.1 GL888184 EGI65621.1 GEDV01006046 JAP82511.1 AAZX01011334 KQ760827 OAD58896.1 GGMR01011522 MBY24141.1 KZ288271 PBC29856.1 HACA01002248 CDW19609.1 ABLF02019741 RCHS01002450 RMX47262.1 GDIQ01213328 JAK38397.1 GDIQ01033043 JAN61694.1 GDIQ01030598 JAN64139.1 GDIQ01168444 JAK83281.1 GDIQ01063281 JAN31456.1 GDIQ01211031 JAK40694.1 GDIQ01211030 JAK40695.1 GDIQ01105516 JAL46210.1 GDIQ01105515 JAL46211.1 GDIQ01016243 JAN78494.1 GDIQ01130108 JAL21618.1 GDIP01019757 JAM83958.1 GDIQ01130109 JAL21617.1 GDIP01035631 JAM68084.1 LRGB01002793 KZS06330.1 GGLE01002088 MBY06214.1
BABH01031276 BABH01031277 AGBW02015068 OWR40641.1 NWSH01000888 PCG73672.1 PCG73674.1 KQ459605 KPI91995.1 KQ461196 KPJ06307.1 ADTU01000400 KQ435706 KOX80133.1 GEZM01099854 JAV53140.1 AXCN02000090 APCN01002223 AAAB01008820 EAA05407.5 KQ980937 KYN11280.1 KQ981606 KYN39551.1 AXCM01000231 AXCM01000232 AXCM01000233 KQ983238 KYQ46225.1 KQ434851 KZC08609.1 GBYB01004819 JAG74586.1 LBMM01001707 KMQ95861.1 KQ977978 KYM98381.1 KQ414581 KOC70975.1 DS232219 EDS37790.1 KB631924 ERL87250.1 APGK01002955 KB735045 ENN83457.1 KK852480 KDR22947.1 QOIP01000002 RLU25817.1 GL448708 EFN83982.1 KK107391 EZA51369.1 CH477665 EAT37541.1 APGK01004030 KB735543 ENN83352.1 KQ971372 EFA10287.2 ADMH02002130 ETN58526.1 KQ976509 KYM82708.1 KYB25206.1 NNAY01001682 OXU23234.1 CVRI01000065 CRL05825.1 GL888184 EGI65621.1 GEDV01006046 JAP82511.1 AAZX01011334 KQ760827 OAD58896.1 GGMR01011522 MBY24141.1 KZ288271 PBC29856.1 HACA01002248 CDW19609.1 ABLF02019741 RCHS01002450 RMX47262.1 GDIQ01213328 JAK38397.1 GDIQ01033043 JAN61694.1 GDIQ01030598 JAN64139.1 GDIQ01168444 JAK83281.1 GDIQ01063281 JAN31456.1 GDIQ01211031 JAK40694.1 GDIQ01211030 JAK40695.1 GDIQ01105516 JAL46210.1 GDIQ01105515 JAL46211.1 GDIQ01016243 JAN78494.1 GDIQ01130108 JAL21618.1 GDIP01019757 JAM83958.1 GDIQ01130109 JAL21617.1 GDIP01035631 JAM68084.1 LRGB01002793 KZS06330.1 GGLE01002088 MBY06214.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000005205
+ More
UP000075880 UP000053105 UP000075886 UP000075885 UP000075882 UP000075840 UP000069272 UP000076407 UP000007062 UP000078492 UP000078541 UP000075883 UP000075884 UP000075903 UP000075881 UP000075809 UP000076502 UP000075902 UP000075900 UP000075920 UP000036403 UP000078542 UP000005203 UP000053825 UP000076408 UP000002320 UP000030742 UP000019118 UP000027135 UP000279307 UP000008237 UP000053097 UP000008820 UP000007266 UP000000673 UP000078540 UP000215335 UP000183832 UP000007755 UP000002358 UP000242457 UP000007819 UP000275408 UP000076858
UP000075880 UP000053105 UP000075886 UP000075885 UP000075882 UP000075840 UP000069272 UP000076407 UP000007062 UP000078492 UP000078541 UP000075883 UP000075884 UP000075903 UP000075881 UP000075809 UP000076502 UP000075902 UP000075900 UP000075920 UP000036403 UP000078542 UP000005203 UP000053825 UP000076408 UP000002320 UP000030742 UP000019118 UP000027135 UP000279307 UP000008237 UP000053097 UP000008820 UP000007266 UP000000673 UP000078540 UP000215335 UP000183832 UP000007755 UP000002358 UP000242457 UP000007819 UP000275408 UP000076858
PRIDE
Interpro
ProteinModelPortal
H9J2W5
A0A212EGN6
A0A2A4JQH4
A0A2A4JQ03
A0A194PLL2
A0A194QL96
+ More
A0A158NQI3 A0A182J7D6 A0A0M9ABX8 A0A1Y1JZC7 A0A182Q1H4 A0A182P8W4 A0A182KX82 A0A182HJY7 A0A182FKZ1 A0A182WW13 Q7QH31 A0A151IUV9 A0A195FGP8 A0A182MEU4 A0A182NL10 A0A182UW75 A0A182JVY1 A0A151WEG8 A0A154P9M1 A0A0C9QMT7 A0A182U8R3 A0A182R877 A0A182VS60 A0A0J7KZS0 A0A151IDA6 A0A088ADA9 A0A0L7RJC9 A0A182YLN0 B0WZW5 U4UBA7 N6UX27 A0A067RGP1 A0A3L8DZI2 E2BJX3 A0A026W8L7 Q16SU8 N6UWR0 D6X2R8 W5J3V5 A0A195BDS5 A0A139WBB9 A0A232EXR6 A0A1J1J4M2 F4WJK5 A0A131YX57 K7JAY2 A0A310SR06 A0A2S2P3S9 A0A2A3EFC8 A0A0K2T0P4 J9JMF7 A0A3M6U0T9 A0A0P5K9L6 A0A0P6GL47 A0A0P6GPY1 A0A0P5LR64 A0A0P6FAD3 A0A0P5IS95 A0A0P5IN93 A0A0P5SEJ8 A0A0N8C843 A0A0P6HP95 A0A0P5PUH7 A0A0P6C7M5 A0A0P5PJ39 A0A0N8DGM7 A0A164NXP5 A0A2R5L9N5
A0A158NQI3 A0A182J7D6 A0A0M9ABX8 A0A1Y1JZC7 A0A182Q1H4 A0A182P8W4 A0A182KX82 A0A182HJY7 A0A182FKZ1 A0A182WW13 Q7QH31 A0A151IUV9 A0A195FGP8 A0A182MEU4 A0A182NL10 A0A182UW75 A0A182JVY1 A0A151WEG8 A0A154P9M1 A0A0C9QMT7 A0A182U8R3 A0A182R877 A0A182VS60 A0A0J7KZS0 A0A151IDA6 A0A088ADA9 A0A0L7RJC9 A0A182YLN0 B0WZW5 U4UBA7 N6UX27 A0A067RGP1 A0A3L8DZI2 E2BJX3 A0A026W8L7 Q16SU8 N6UWR0 D6X2R8 W5J3V5 A0A195BDS5 A0A139WBB9 A0A232EXR6 A0A1J1J4M2 F4WJK5 A0A131YX57 K7JAY2 A0A310SR06 A0A2S2P3S9 A0A2A3EFC8 A0A0K2T0P4 J9JMF7 A0A3M6U0T9 A0A0P5K9L6 A0A0P6GL47 A0A0P6GPY1 A0A0P5LR64 A0A0P6FAD3 A0A0P5IS95 A0A0P5IN93 A0A0P5SEJ8 A0A0N8C843 A0A0P6HP95 A0A0P5PUH7 A0A0P6C7M5 A0A0P5PJ39 A0A0N8DGM7 A0A164NXP5 A0A2R5L9N5
Ontologies
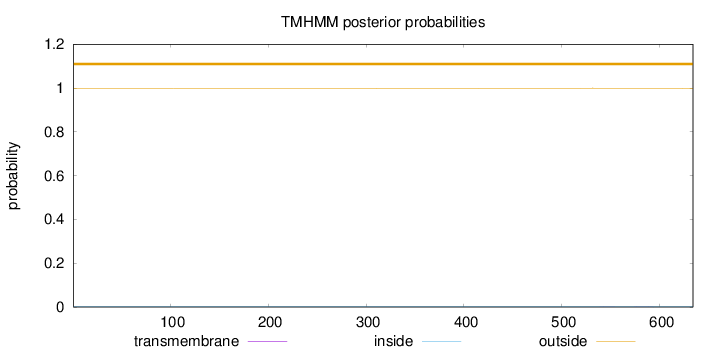
Topology
Length:
634
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00928
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.00059
outside
1 - 634
Population Genetic Test Statistics
Pi
16.911976
Theta
2.361996
Tajima's D
-0.956883
CLR
0.004295
CSRT
0.144792760361982
Interpretation
Uncertain
