Gene
KWMTBOMO00617
Pre Gene Modal
BGIBMGA003900
Annotation
PREDICTED:_protein_angel-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.865
Sequence
CDS
ATGCTGAAGACAAACAGTTTTTGCAGATTATTGAAAATAGTCACAGACAACACATATTGCGGGGCACATGTCAGGAAGTTTGCATATTTAAGCAGGAAGAAGTCATGCTTGCCTCAGTGCTTGGCGAGAGCTACGTGTTTGCACACGGGTCTGCTGCTGCGACAGAAGAAACGGTTGAGGATAAAGATACCAACATTTCACTCCGGTGCGAGTACGACAAAAGGGCAAATGAGTGACGAGAGCCAGCCTACCGACAATCATGCCAGTTTCGGAGGAACTTTTGAAGTTAGTGATTCATCCGGCGAAGACACGTCGATTGTTGAACAGAAATATGATTCTGAGGACGCGAAATCCGAGCACCACAGGAGACCAACAAAGATCCCACTCAACTTCAGGACCTGGGAGACTGTAGGGAAGAAGACCGACTCGGACGAGACAACGTTTAGGTTCAAAGTGGTGTCTTACAATGTACTAGCGCAGTATTTGTTGGAGTATCATCCGTATCTCTACATTGATTGCTCTCCGAGGAACTTAAAGTGGAAACACAGATCTAGACGTCTCTACCAAGAAATTCGCAGACTGTCACCTGACATACTGTGCCTCCAAGAAGTTCAACTGTCGCACCTTGAAACGTTCTATTCTAAATTCGAAAACATCGGGTACCAAGGTGTCTTCAAACAGAAAACCGGAGACAGGCAGGACGGATGCGCTATTTACTTCAAGAAGTCTTTATTTGATTTGGACGATCAAATCAGCGTTGAGTTCTTCCAGCCGGAGCTGCCTATCCTCAACCGTGATAATATCGGTGTGATTGTTAAATTGTCGCCGAAGGAATTGCCCGGGTCTTCCATAGTCGTTGCCACAACTCACTTACTCTACAATCCGAAGAGAACGGACGTACGACTAGCACAAATAAAAATATTACTCGCCGAGATCGATAGATTCGCTTATTTCAACAATGGGAGGGAGTCCGGCCATTTTCCCGTTATACTGACAGGCGATTTCAACTCCACCCCAGAAAGTGCTGTGATAAAATTACTAGATAGAGGACGCGTTAGAGCGAGTTCGCTACGAGACAATTCGGATTGGAAACGAATCGGCGTCACCGATAACTGCCAGCACTTGGCCGTTTATTTAAACCGACAACAGGGATGTCCACCGAATTTCAACAAAATTAAGATACACAACTCGGAATACGCTTTCGACTCGGCCGACGCGGACGAGAGTGATTACTGGAACGATGCTCACAATCGGATGTTCAACAGTGACGTCATAGGTCACTCCCTAAACCTGATCTCTGTTTACAACAAGTTCAAGGCGGACGGGCAAATGGAGGCCTCCACTTTCCAAGACCATTGGGTCACGGTCGATTACATTTATTTTAGCTGTCGCAGCGGTCTCCGTCTGCTCGAGCGTCTGCGGCTGCCGACGGCCGCGGAGTGCGGCGTGCTCGGTACGCTCCCGAACGAAGTGTACCCCTCGGACCATCTGGCGACCGCGGCCGTGTTCGAGCTGAGGCGGTTCCGCTACGCCTCCTAA
Protein
MLKTNSFCRLLKIVTDNTYCGAHVRKFAYLSRKKSCLPQCLARATCLHTGLLLRQKKRLRIKIPTFHSGASTTKGQMSDESQPTDNHASFGGTFEVSDSSGEDTSIVEQKYDSEDAKSEHHRRPTKIPLNFRTWETVGKKTDSDETTFRFKVVSYNVLAQYLLEYHPYLYIDCSPRNLKWKHRSRRLYQEIRRLSPDILCLQEVQLSHLETFYSKFENIGYQGVFKQKTGDRQDGCAIYFKKSLFDLDDQISVEFFQPELPILNRDNIGVIVKLSPKELPGSSIVVATTHLLYNPKRTDVRLAQIKILLAEIDRFAYFNNGRESGHFPVILTGDFNSTPESAVIKLLDRGRVRASSLRDNSDWKRIGVTDNCQHLAVYLNRQQGCPPNFNKIKIHNSEYAFDSADADESDYWNDAHNRMFNSDVIGHSLNLISVYNKFKADGQMEASTFQDHWVTVDYIYFSCRSGLRLLERLRLPTAAECGVLGTLPNEVYPSDHLATAAVFELRRFRYAS
Summary
Uniprot
A0A2A4JIB6
A0A212EGP1
A0A2H1V0B4
A0A194PF50
A0A194QRY8
A0A0L7KZR6
+ More
A0A2A4K1Z1 A0A2H1WMX3 A0A336L0E7 A0A336N0R2 D2A3E6 A0A151WP77 A0A1B6LEP8 E2BU60 E2AGN8 N6UA56 A0A0M8ZYL9 A0A1B6GP70 A0A1B6F7X5 A0A195DNV8 A0A1B6FX57 F4W625 A0A158P1V2 A0A195EQE3 A0A195AWK0 A0A151K2A1 A0A1Y1L5P2 A0A1Y1L9K0 E9J3S8 A0A1Y1L5P1 A0A0L7QK42 A0A194QKP4 A0A1Y1L868 A0A232F4Y3 A0A0P6FIL6 K7J539 A0A0N8E5Q7 A0A1W4WIV4 A0A1S4F2X5 Q17H00 A0A182GQX0 A0A0P6G2F1 A0A0P5Y1R8 A0A0P6FJ83 A0A0P5T407 A0A0N8BW34 A0A164T0Z4 A0A0P5DCA2 A0A0P6CQ58 A0A0N8EKA4 A0A0P4ZF73 A0A026WKF7 A0A182MCT7 A0A067RFP6 E9I279 A0A069DYQ9 U5ES81 A0A0P6I0P4 A0A0P5IVT5 A0A3L8E108 A0A0C9R0Y4 A0A2J7RKV8 C4WV61 A0A224XEY4 A0A0P5ISC2 A0A182TPY6 A0A182NPK7 A0A182WG27 A0A0P4VI84 A0A0V0G9T0 A0A1Q3FEE3 A0A0P5FZU4 A0A1Q3FEB9 A0A182YD93 E0W1J1 A0A2S2QM84 A0A084WB29 A0A0P5LW01 A0A182QN36 A0A154PJ15 A0A2J7RKW2 W5JHE6 A0A2H8TGX3 A0A182FBT1 A0A182RB77 B0W7B1 A0A0P6DL17 A0A0P6F996 A0A0P5URK7 A0A0N8ED62 A0A0P5VMU3 A0A2M4BNX2 A0A182XEC8 A0A0A9XHZ2 A0A2M4BPR9 A0A0A9XT95 A0A182JVB5
A0A2A4K1Z1 A0A2H1WMX3 A0A336L0E7 A0A336N0R2 D2A3E6 A0A151WP77 A0A1B6LEP8 E2BU60 E2AGN8 N6UA56 A0A0M8ZYL9 A0A1B6GP70 A0A1B6F7X5 A0A195DNV8 A0A1B6FX57 F4W625 A0A158P1V2 A0A195EQE3 A0A195AWK0 A0A151K2A1 A0A1Y1L5P2 A0A1Y1L9K0 E9J3S8 A0A1Y1L5P1 A0A0L7QK42 A0A194QKP4 A0A1Y1L868 A0A232F4Y3 A0A0P6FIL6 K7J539 A0A0N8E5Q7 A0A1W4WIV4 A0A1S4F2X5 Q17H00 A0A182GQX0 A0A0P6G2F1 A0A0P5Y1R8 A0A0P6FJ83 A0A0P5T407 A0A0N8BW34 A0A164T0Z4 A0A0P5DCA2 A0A0P6CQ58 A0A0N8EKA4 A0A0P4ZF73 A0A026WKF7 A0A182MCT7 A0A067RFP6 E9I279 A0A069DYQ9 U5ES81 A0A0P6I0P4 A0A0P5IVT5 A0A3L8E108 A0A0C9R0Y4 A0A2J7RKV8 C4WV61 A0A224XEY4 A0A0P5ISC2 A0A182TPY6 A0A182NPK7 A0A182WG27 A0A0P4VI84 A0A0V0G9T0 A0A1Q3FEE3 A0A0P5FZU4 A0A1Q3FEB9 A0A182YD93 E0W1J1 A0A2S2QM84 A0A084WB29 A0A0P5LW01 A0A182QN36 A0A154PJ15 A0A2J7RKW2 W5JHE6 A0A2H8TGX3 A0A182FBT1 A0A182RB77 B0W7B1 A0A0P6DL17 A0A0P6F996 A0A0P5URK7 A0A0N8ED62 A0A0P5VMU3 A0A2M4BNX2 A0A182XEC8 A0A0A9XHZ2 A0A2M4BPR9 A0A0A9XT95 A0A182JVB5
Pubmed
EMBL
NWSH01001383
PCG71458.1
AGBW02015068
OWR40640.1
ODYU01000091
SOQ34277.1
+ More
KQ459605 KPI91996.1 KQ461196 KPJ06306.1 JTDY01003976 KOB68753.1 NWSH01000282 PCG77783.1 ODYU01009682 SOQ54282.1 UFQS01001373 UFQT01001373 SSX10560.1 SSX30244.1 UFQT01002915 SSX34277.1 KQ971338 EFA02304.2 KQ982892 KYQ49608.1 GEBQ01017826 JAT22151.1 GL450581 EFN80777.1 GL439357 EFN67403.1 APGK01033055 KB740860 KB632287 ENN78580.1 ERL91464.1 KQ435824 KOX72239.1 GECZ01005616 JAS64153.1 GECZ01023442 JAS46327.1 KQ980724 KYN14169.1 GECZ01015146 JAS54623.1 GL887707 EGI70160.1 ADTU01006889 KQ982021 KYN30433.1 KQ976731 KYM76344.1 LKEX01012869 KYN50202.1 GEZM01063724 JAV68993.1 GEZM01063727 JAV68990.1 GL768069 EFZ12453.1 GEZM01063725 JAV68992.1 KQ415041 KOC58886.1 KQ461198 KPJ06133.1 GEZM01063726 JAV68991.1 NNAY01000936 OXU25841.1 GDIQ01059213 JAN35524.1 AAZX01000348 GDIQ01059212 JAN35525.1 CH477254 EAT45924.1 JXUM01081564 JXUM01081565 KQ563255 KXJ74189.1 GDIQ01050518 JAN44219.1 GDIP01065205 JAM38510.1 GDIQ01056566 JAN38171.1 GDIP01135796 JAL67918.1 GDIQ01139187 GDIQ01048419 JAL12539.1 LRGB01001880 KZS10121.1 GDIP01158189 JAJ65213.1 GDIP01010980 JAM92735.1 GDIQ01018436 JAN76301.1 GDIP01214182 JAJ09220.1 KK107163 EZA56537.1 AXCM01002291 KK852498 KDR22666.1 GL734001 EFX61903.1 GBGD01001960 JAC86929.1 GANO01003368 JAB56503.1 GDIQ01036318 JAN58419.1 GDIQ01208360 JAK43365.1 QOIP01000002 RLU25708.1 GBYB01001649 GBYB01001650 JAG71416.1 JAG71417.1 NEVH01002706 PNF41478.1 AK341372 BAH71781.1 GFTR01005390 JAW11036.1 GDIQ01209748 JAK41977.1 GDKW01003074 JAI53521.1 GECL01002103 JAP04021.1 GFDL01009118 JAV25927.1 GDIQ01255125 JAJ96599.1 GFDL01009156 JAV25889.1 DS235870 EEB19497.1 GGMS01009598 MBY78801.1 ATLV01022303 KE525331 KFB47423.1 GDIQ01167264 JAK84461.1 AXCN02002107 KQ434916 KZC11444.1 PNF41477.1 ADMH02001184 ETN63792.1 GFXV01001540 MBW13345.1 DS231853 EDS37826.1 GDIQ01090801 JAN03936.1 GDIQ01053618 JAN41119.1 GDIP01126592 JAL77122.1 GDIQ01038372 JAN56365.1 GDIP01097905 JAM05810.1 GGFJ01005639 MBW54780.1 GBHO01023257 JAG20347.1 GGFJ01005637 MBW54778.1 GBHO01023258 GBHO01023254 JAG20346.1 JAG20350.1
KQ459605 KPI91996.1 KQ461196 KPJ06306.1 JTDY01003976 KOB68753.1 NWSH01000282 PCG77783.1 ODYU01009682 SOQ54282.1 UFQS01001373 UFQT01001373 SSX10560.1 SSX30244.1 UFQT01002915 SSX34277.1 KQ971338 EFA02304.2 KQ982892 KYQ49608.1 GEBQ01017826 JAT22151.1 GL450581 EFN80777.1 GL439357 EFN67403.1 APGK01033055 KB740860 KB632287 ENN78580.1 ERL91464.1 KQ435824 KOX72239.1 GECZ01005616 JAS64153.1 GECZ01023442 JAS46327.1 KQ980724 KYN14169.1 GECZ01015146 JAS54623.1 GL887707 EGI70160.1 ADTU01006889 KQ982021 KYN30433.1 KQ976731 KYM76344.1 LKEX01012869 KYN50202.1 GEZM01063724 JAV68993.1 GEZM01063727 JAV68990.1 GL768069 EFZ12453.1 GEZM01063725 JAV68992.1 KQ415041 KOC58886.1 KQ461198 KPJ06133.1 GEZM01063726 JAV68991.1 NNAY01000936 OXU25841.1 GDIQ01059213 JAN35524.1 AAZX01000348 GDIQ01059212 JAN35525.1 CH477254 EAT45924.1 JXUM01081564 JXUM01081565 KQ563255 KXJ74189.1 GDIQ01050518 JAN44219.1 GDIP01065205 JAM38510.1 GDIQ01056566 JAN38171.1 GDIP01135796 JAL67918.1 GDIQ01139187 GDIQ01048419 JAL12539.1 LRGB01001880 KZS10121.1 GDIP01158189 JAJ65213.1 GDIP01010980 JAM92735.1 GDIQ01018436 JAN76301.1 GDIP01214182 JAJ09220.1 KK107163 EZA56537.1 AXCM01002291 KK852498 KDR22666.1 GL734001 EFX61903.1 GBGD01001960 JAC86929.1 GANO01003368 JAB56503.1 GDIQ01036318 JAN58419.1 GDIQ01208360 JAK43365.1 QOIP01000002 RLU25708.1 GBYB01001649 GBYB01001650 JAG71416.1 JAG71417.1 NEVH01002706 PNF41478.1 AK341372 BAH71781.1 GFTR01005390 JAW11036.1 GDIQ01209748 JAK41977.1 GDKW01003074 JAI53521.1 GECL01002103 JAP04021.1 GFDL01009118 JAV25927.1 GDIQ01255125 JAJ96599.1 GFDL01009156 JAV25889.1 DS235870 EEB19497.1 GGMS01009598 MBY78801.1 ATLV01022303 KE525331 KFB47423.1 GDIQ01167264 JAK84461.1 AXCN02002107 KQ434916 KZC11444.1 PNF41477.1 ADMH02001184 ETN63792.1 GFXV01001540 MBW13345.1 DS231853 EDS37826.1 GDIQ01090801 JAN03936.1 GDIQ01053618 JAN41119.1 GDIP01126592 JAL77122.1 GDIQ01038372 JAN56365.1 GDIP01097905 JAM05810.1 GGFJ01005639 MBW54780.1 GBHO01023257 JAG20347.1 GGFJ01005637 MBW54778.1 GBHO01023258 GBHO01023254 JAG20346.1 JAG20350.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
UP000007266
+ More
UP000075809 UP000008237 UP000000311 UP000019118 UP000030742 UP000053105 UP000078492 UP000007755 UP000005205 UP000078541 UP000078540 UP000078542 UP000053825 UP000215335 UP000002358 UP000192223 UP000008820 UP000069940 UP000249989 UP000076858 UP000053097 UP000075883 UP000027135 UP000000305 UP000279307 UP000235965 UP000075902 UP000075884 UP000075920 UP000076408 UP000009046 UP000030765 UP000075886 UP000076502 UP000000673 UP000069272 UP000075900 UP000002320 UP000076407 UP000075881
UP000075809 UP000008237 UP000000311 UP000019118 UP000030742 UP000053105 UP000078492 UP000007755 UP000005205 UP000078541 UP000078540 UP000078542 UP000053825 UP000215335 UP000002358 UP000192223 UP000008820 UP000069940 UP000249989 UP000076858 UP000053097 UP000075883 UP000027135 UP000000305 UP000279307 UP000235965 UP000075902 UP000075884 UP000075920 UP000076408 UP000009046 UP000030765 UP000075886 UP000076502 UP000000673 UP000069272 UP000075900 UP000002320 UP000076407 UP000075881
Pfam
PF03372 Exo_endo_phos
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
A0A2A4JIB6
A0A212EGP1
A0A2H1V0B4
A0A194PF50
A0A194QRY8
A0A0L7KZR6
+ More
A0A2A4K1Z1 A0A2H1WMX3 A0A336L0E7 A0A336N0R2 D2A3E6 A0A151WP77 A0A1B6LEP8 E2BU60 E2AGN8 N6UA56 A0A0M8ZYL9 A0A1B6GP70 A0A1B6F7X5 A0A195DNV8 A0A1B6FX57 F4W625 A0A158P1V2 A0A195EQE3 A0A195AWK0 A0A151K2A1 A0A1Y1L5P2 A0A1Y1L9K0 E9J3S8 A0A1Y1L5P1 A0A0L7QK42 A0A194QKP4 A0A1Y1L868 A0A232F4Y3 A0A0P6FIL6 K7J539 A0A0N8E5Q7 A0A1W4WIV4 A0A1S4F2X5 Q17H00 A0A182GQX0 A0A0P6G2F1 A0A0P5Y1R8 A0A0P6FJ83 A0A0P5T407 A0A0N8BW34 A0A164T0Z4 A0A0P5DCA2 A0A0P6CQ58 A0A0N8EKA4 A0A0P4ZF73 A0A026WKF7 A0A182MCT7 A0A067RFP6 E9I279 A0A069DYQ9 U5ES81 A0A0P6I0P4 A0A0P5IVT5 A0A3L8E108 A0A0C9R0Y4 A0A2J7RKV8 C4WV61 A0A224XEY4 A0A0P5ISC2 A0A182TPY6 A0A182NPK7 A0A182WG27 A0A0P4VI84 A0A0V0G9T0 A0A1Q3FEE3 A0A0P5FZU4 A0A1Q3FEB9 A0A182YD93 E0W1J1 A0A2S2QM84 A0A084WB29 A0A0P5LW01 A0A182QN36 A0A154PJ15 A0A2J7RKW2 W5JHE6 A0A2H8TGX3 A0A182FBT1 A0A182RB77 B0W7B1 A0A0P6DL17 A0A0P6F996 A0A0P5URK7 A0A0N8ED62 A0A0P5VMU3 A0A2M4BNX2 A0A182XEC8 A0A0A9XHZ2 A0A2M4BPR9 A0A0A9XT95 A0A182JVB5
A0A2A4K1Z1 A0A2H1WMX3 A0A336L0E7 A0A336N0R2 D2A3E6 A0A151WP77 A0A1B6LEP8 E2BU60 E2AGN8 N6UA56 A0A0M8ZYL9 A0A1B6GP70 A0A1B6F7X5 A0A195DNV8 A0A1B6FX57 F4W625 A0A158P1V2 A0A195EQE3 A0A195AWK0 A0A151K2A1 A0A1Y1L5P2 A0A1Y1L9K0 E9J3S8 A0A1Y1L5P1 A0A0L7QK42 A0A194QKP4 A0A1Y1L868 A0A232F4Y3 A0A0P6FIL6 K7J539 A0A0N8E5Q7 A0A1W4WIV4 A0A1S4F2X5 Q17H00 A0A182GQX0 A0A0P6G2F1 A0A0P5Y1R8 A0A0P6FJ83 A0A0P5T407 A0A0N8BW34 A0A164T0Z4 A0A0P5DCA2 A0A0P6CQ58 A0A0N8EKA4 A0A0P4ZF73 A0A026WKF7 A0A182MCT7 A0A067RFP6 E9I279 A0A069DYQ9 U5ES81 A0A0P6I0P4 A0A0P5IVT5 A0A3L8E108 A0A0C9R0Y4 A0A2J7RKV8 C4WV61 A0A224XEY4 A0A0P5ISC2 A0A182TPY6 A0A182NPK7 A0A182WG27 A0A0P4VI84 A0A0V0G9T0 A0A1Q3FEE3 A0A0P5FZU4 A0A1Q3FEB9 A0A182YD93 E0W1J1 A0A2S2QM84 A0A084WB29 A0A0P5LW01 A0A182QN36 A0A154PJ15 A0A2J7RKW2 W5JHE6 A0A2H8TGX3 A0A182FBT1 A0A182RB77 B0W7B1 A0A0P6DL17 A0A0P6F996 A0A0P5URK7 A0A0N8ED62 A0A0P5VMU3 A0A2M4BNX2 A0A182XEC8 A0A0A9XHZ2 A0A2M4BPR9 A0A0A9XT95 A0A182JVB5
PDB
4B8C
E-value=4.7668e-23,
Score=268
Ontologies
Topology
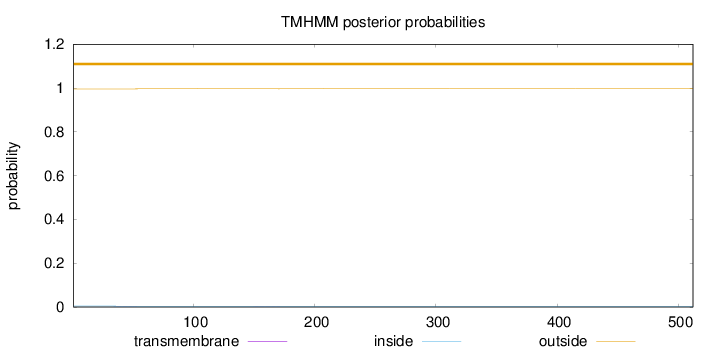
Length:
512
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0180499999999999
Exp number, first 60 AAs:
0.00833
Total prob of N-in:
0.00417
outside
1 - 512
Population Genetic Test Statistics
Pi
19.811791
Theta
22.928645
Tajima's D
-1.1926
CLR
0.001561
CSRT
0.105894705264737
Interpretation
Uncertain
