Gene
KWMTBOMO00614
Pre Gene Modal
BGIBMGA003851
Annotation
PREDICTED:_potassium_voltage-gated_channel_protein_Shaker_isoform_X11_[Bombyx_mori]
Full name
Potassium voltage-gated channel protein Shaker
Alternative Name
Protein minisleep
Location in the cell
PlasmaMembrane Reliability : 4.761
Sequence
CDS
ATGGTGAGCGGTTTGAAGTTCGAAACTCAGCTGCGTACATTGAACCAATTCCCGGAGACGTTGCTCGGGGACCCCTCCCGCCGAATACGATACTTCGACCCACTCCGGAACGAGTACTTCTTCGACCGGAACCGACCGTCTTTCGACGCCATCCTCTACTATTACCAAAGCGGAGGACGACTCCGTCGCCCGGTAAACGTCCCCTTGGACGTCTTTTCTGAAGAGATCAAGTTCTACGAACTGGGAGAGCAGGCCACCAACAAGTTCCGTGAGGATGAAGGTTTCATCAAGGAGGAAGAGAAGCCCCTTCCCTCCAACGAACGCCAGCGCAAAATCTGGCTGCTCTTCGAATACCCCGAGAGCTCCCAGGCCGCGCGCGTCGTTGCCATCATCTCGGTCTTCGTCATCCTCCTCTCCATCGTCATCTTCTGCCTGGAGACCCTTCCTGAATTCAAACACTACAAAGTCTTCAATACAACGACCAACGGCACGAAGATTGAGGAGGACGAGGTCCCGGATATCACCGATCCATTCTTCCTTATTGAAACACTCTGCATCATCTGGTTCACCTTCGAATTGATCGTGCGGTTCTTGGCGTGTCCCAACAAATTCAACTTCTTCAGAGACGTCATGAACATCATTGATATAATCGCCATTATCCCGTACTTCATCACCCTGGCGACCGTCGTTGCCGAGGAGGAAGACACTTTGAACCTACCGCGAGCTCCCGTATCTCCGCAAGATAAGTCCACCAACCAAGCCATGAGCCTGGCTATTCTGCGGGTAATCCGTCTCGTGCGGGTGTTCCGGATCTTCAAACTGTCCCGCCACTCGAAGGGATTGCAGATTCTTGGCAGGACGCTCAAGGCGTCCATGCGAGAACTTGGCCTGCTGATCTTCTTCCTGTTCATCGGTGTGGTGCTGTTTTCGTCTGCGGTGTACTTCGCTGAAGCCGGAAGCGAGAACAGCTTCTTCAAATCCATACCTGATGCGTTTTGGTGGGCGGTCGTGACAATGACGACCGTAGGATACGGAGACATGACGCCGGTCGGAGTCTGGGGGAAAATCGTGGGTTCGCTGTGCGCCATCGCCGGCGTGCTCACTATCGCCCTCCCCGTGCCCGTCATCGTCTCCAACTTCAACTACTTCTACCACCGCGAAACTGACCAGGAAGAGATGCAGTCCCAGAACTTCAACCACGTAACCAGCTGCCCCTACCTGCCGGGAACCATGGGCGAGCCCTACCTGATTTCAGGGCCAATACAGAAGAAGTCCTCTATGAGCGATGACTCCTCTTCTGAGATGATGGAGTTGGACGAGTCGGCCATGGCGTCCGAGCCGCAACTCGAGCAGAACCTGCGCCGCCTCCGCGAGGAGGAGAACCTTCTGCGCGAGCAGCAAGCCACCATGGAGGCCGGTGCGTTCGACGACATCGCCGCTCTGGAGCATCAGGACGCTCTGCGCCTCAACCAGGTTACTCATCTTTTTTACGTAATATCTTCATTAACCATTTACGGATACTAA
Protein
MVSGLKFETQLRTLNQFPETLLGDPSRRIRYFDPLRNEYFFDRNRPSFDAILYYYQSGGRLRRPVNVPLDVFSEEIKFYELGEQATNKFREDEGFIKEEEKPLPSNERQRKIWLLFEYPESSQAARVVAIISVFVILLSIVIFCLETLPEFKHYKVFNTTTNGTKIEEDEVPDITDPFFLIETLCIIWFTFELIVRFLACPNKFNFFRDVMNIIDIIAIIPYFITLATVVAEEEDTLNLPRAPVSPQDKSTNQAMSLAILRVIRLVRVFRIFKLSRHSKGLQILGRTLKASMRELGLLIFFLFIGVVLFSSAVYFAEAGSENSFFKSIPDAFWWAVVTMTTVGYGDMTPVGVWGKIVGSLCAIAGVLTIALPVPVIVSNFNYFYHRETDQEEMQSQNFNHVTSCPYLPGTMGEPYLISGPIQKKSSMSDDSSSEMMELDESAMASEPQLEQNLRRLREEENLLREQQATMEAGAFDDIAALEHQDALRLNQVTHLFYVISSLTIYGY
Summary
Description
Voltage-gated potassium channel that mediates transmembrane potassium transport in excitable membranes. The channel alternates between opened and closed conformations in response to the voltage difference across the membrane. Forms rapidly inactivating tetrameric potassium-selective channels through which potassium ions pass in accordance with their electrochemical gradient and may contribute to A-type currents (PubMed:2448636). Plays a role in the regulation of sleep need or efficiency (PubMed:15858564).
Subunit
Homotetramer or heterotetramer of potassium channel proteins.
Similarity
Belongs to the potassium channel family.
Belongs to the potassium channel family. A (Shaker) (TC 1.A.1.2) subfamily. Shaker sub-subfamily.
Belongs to the potassium channel family. A (Shaker) (TC 1.A.1.2) subfamily. Shaker sub-subfamily.
Keywords
3D-structure
Alternative splicing
Cell membrane
Complete proteome
Glycoprotein
Ion channel
Ion transport
Membrane
Potassium
Potassium channel
Potassium transport
Reference proteome
RNA editing
Transmembrane
Transmembrane helix
Transport
Voltage-gated channel
Feature
chain Potassium voltage-gated channel protein Shaker
splice variant In isoform Beta and isoform L.
splice variant In isoform Beta and isoform L.
Uniprot
H9J2W4
E9LP24
A0A194PLL7
A0A194QL53
A0A2A4JHA3
A0A2M4ASP2
+ More
A0A2H1V1X8 A0A1S4EVA2 A0A348G660 A0A2M4ALN5 Q7QF14 Q17PT9 A0A0P8ZSJ5 B4NPI1 A0A1W4UMX6 B4I6L8 B3NWZ6 B4Q2M4 P08510 B4JJ88 A0A1W4UNF5 B4L2A7 B4M6S3 A0A182QK67 A0A0R1EDM7 A0A0Q9W6W6 T1HFA1 P08510-3 A0A1W4V0Q2 A0A0Q9W709 A0A0Q9X091 A0A0Q5T428 A0A0R1EIB4 A0A1W4UNG0 A0A0Q9WI52 A0A1I8MXT4 A0A1W4V1X9 A0A2S1ZNM0 A0A1W4V0Q7 A0A0Q9WEM3 A0A0Q9WYW2 A0A0Q9WNB8 A0A0Q9WPL7 A0A1W4WTF4 A0A1W4UMY0 A0A1W4V0P7 W8AV96 A0A1W4WUD3 A0A0Q9W771 A0A0Q9WN85 A0A1W4X4N1 F4WVX1 A0A1I8Q8D2 A0A026WWX1 F5HJN5 P08510-9 A0A1W4X4K0 A0A1W4X443 A0A1W4V0D3 A0A0Q9WVC7 A0A1W4X2C8 W8AJ67 A0A1I8MXT7 A0A151I5G4 A0A3L8D818 A0A151WRY2 A0A1I8Q8G9 A0A1I8Q8H1 A0A0S0X7Z4 A0A1I8Q8D6 A0A1W4UMY5 A0A195DUN5 D6WHV5 P08510-7 A0A1Y1L627 A0A0R1EC30 A0A151K1K0 A0A1W4V0D8 A0A1W4UNG5 A0A0R1EIL7 A0A0N8P0L6 A0A0P8Y7Q9 A0A0Q5T3U2 A0A0Q5TEG4 E2AQU3 A0A0Q9WNA1 A0A0R1EBV3 P08510-2 A0A224XNR1 A0A0R1ECH3 A0A0R1EC16 A0A0P9C1E9 A0A0P9AI13 A0A0Q5T3T6 A0A0Q5T3R9 A0A0Q5T6N6 A0A0Q5TJD7 A0A0P8XS21 A0A0R1EFN3 A0A0R1EJQ1
A0A2H1V1X8 A0A1S4EVA2 A0A348G660 A0A2M4ALN5 Q7QF14 Q17PT9 A0A0P8ZSJ5 B4NPI1 A0A1W4UMX6 B4I6L8 B3NWZ6 B4Q2M4 P08510 B4JJ88 A0A1W4UNF5 B4L2A7 B4M6S3 A0A182QK67 A0A0R1EDM7 A0A0Q9W6W6 T1HFA1 P08510-3 A0A1W4V0Q2 A0A0Q9W709 A0A0Q9X091 A0A0Q5T428 A0A0R1EIB4 A0A1W4UNG0 A0A0Q9WI52 A0A1I8MXT4 A0A1W4V1X9 A0A2S1ZNM0 A0A1W4V0Q7 A0A0Q9WEM3 A0A0Q9WYW2 A0A0Q9WNB8 A0A0Q9WPL7 A0A1W4WTF4 A0A1W4UMY0 A0A1W4V0P7 W8AV96 A0A1W4WUD3 A0A0Q9W771 A0A0Q9WN85 A0A1W4X4N1 F4WVX1 A0A1I8Q8D2 A0A026WWX1 F5HJN5 P08510-9 A0A1W4X4K0 A0A1W4X443 A0A1W4V0D3 A0A0Q9WVC7 A0A1W4X2C8 W8AJ67 A0A1I8MXT7 A0A151I5G4 A0A3L8D818 A0A151WRY2 A0A1I8Q8G9 A0A1I8Q8H1 A0A0S0X7Z4 A0A1I8Q8D6 A0A1W4UMY5 A0A195DUN5 D6WHV5 P08510-7 A0A1Y1L627 A0A0R1EC30 A0A151K1K0 A0A1W4V0D8 A0A1W4UNG5 A0A0R1EIL7 A0A0N8P0L6 A0A0P8Y7Q9 A0A0Q5T3U2 A0A0Q5TEG4 E2AQU3 A0A0Q9WNA1 A0A0R1EBV3 P08510-2 A0A224XNR1 A0A0R1ECH3 A0A0R1EC16 A0A0P9C1E9 A0A0P9AI13 A0A0Q5T3T6 A0A0Q5T3R9 A0A0Q5T6N6 A0A0Q5TJD7 A0A0P8XS21 A0A0R1EFN3 A0A0R1EJQ1
Pubmed
19121390
21672063
26354079
12364791
17510324
17994087
+ More
18057021 17550304 2441471 16453805 2456921 2448635 10731132 12537572 12537569 2440582 3272175 2448636 12907802 15858564 17893096 12023222 25315136 24495485 21719571 24508170 30249741 12537568 12537573 12537574 16110336 17569856 17569867 18362917 19820115 28004739 20798317
18057021 17550304 2441471 16453805 2456921 2448635 10731132 12537572 12537569 2440582 3272175 2448636 12907802 15858564 17893096 12023222 25315136 24495485 21719571 24508170 30249741 12537568 12537573 12537574 16110336 17569856 17569867 18362917 19820115 28004739 20798317
EMBL
BABH01031285
HQ116693
ADV76532.1
KQ459605
KPI92000.1
KQ461196
+ More
KPJ06303.1 NWSH01001383 PCG71455.1 GGFK01010472 MBW43793.1 ODYU01000091 SOQ34274.1 FX985601 BBF97933.1 GGFK01008369 MBW41690.1 AAAB01008846 EAA06439.5 CH477189 EAT48752.1 CH902630 KPU77466.1 CH964291 EDW86421.1 CH480823 EDW55966.1 CH954180 EDV46543.1 KQS30015.1 KQS30026.1 CM000162 EDX02665.1 KRK06900.1 KRK06902.1 KRK06905.1 M17211 X06184 X07131 X07132 X07133 X07134 X06742 AE014298 AY118398 BT050432 M17155 X78908 CH916370 EDV99640.1 CH933810 EDW07768.1 CH940653 EDW62490.1 KRF80680.1 KRF80687.1 AXCN02002242 AXCN02002243 AXCN02002244 KRK06894.1 KRF80682.1 ACPB03010323 KRF80684.1 KRF94202.1 KQS30029.1 KRK06892.1 KRK06898.1 KRK06904.1 KRF80686.1 MF359241 AWK26944.1 KRF80679.1 KRF94201.1 KRF94200.1 KRF94203.1 GAMC01017997 JAB88558.1 KRF80685.1 KRF94206.1 GL888402 EGI61652.1 KK107078 EZA60221.1 EGK96496.1 KRF94204.1 GAMC01017995 JAB88560.1 KQ976416 KYM90057.1 QOIP01000011 RLU16640.1 KQ982803 KYQ50557.1 ALI51156.1 KQ980322 KYN16561.1 KQ971321 EFA00701.2 GEZM01066536 JAV67820.1 KRK06909.1 KQ981236 KYN44316.1 KRK06906.1 KPU77468.1 KPU77467.1 KQS30028.1 KQS30021.1 GL441834 EFN64205.1 KRF94205.1 KRF94207.1 KRK06895.1 KRK06907.1 GFTR01006797 JAW09629.1 KRK06901.1 KRK06893.1 KPU77465.1 KPU77470.1 KQS30025.1 KQS30019.1 KQS30024.1 KQS30022.1 KPU77469.1 KPU77473.1 KRK06899.1 KRK06903.1
KPJ06303.1 NWSH01001383 PCG71455.1 GGFK01010472 MBW43793.1 ODYU01000091 SOQ34274.1 FX985601 BBF97933.1 GGFK01008369 MBW41690.1 AAAB01008846 EAA06439.5 CH477189 EAT48752.1 CH902630 KPU77466.1 CH964291 EDW86421.1 CH480823 EDW55966.1 CH954180 EDV46543.1 KQS30015.1 KQS30026.1 CM000162 EDX02665.1 KRK06900.1 KRK06902.1 KRK06905.1 M17211 X06184 X07131 X07132 X07133 X07134 X06742 AE014298 AY118398 BT050432 M17155 X78908 CH916370 EDV99640.1 CH933810 EDW07768.1 CH940653 EDW62490.1 KRF80680.1 KRF80687.1 AXCN02002242 AXCN02002243 AXCN02002244 KRK06894.1 KRF80682.1 ACPB03010323 KRF80684.1 KRF94202.1 KQS30029.1 KRK06892.1 KRK06898.1 KRK06904.1 KRF80686.1 MF359241 AWK26944.1 KRF80679.1 KRF94201.1 KRF94200.1 KRF94203.1 GAMC01017997 JAB88558.1 KRF80685.1 KRF94206.1 GL888402 EGI61652.1 KK107078 EZA60221.1 EGK96496.1 KRF94204.1 GAMC01017995 JAB88560.1 KQ976416 KYM90057.1 QOIP01000011 RLU16640.1 KQ982803 KYQ50557.1 ALI51156.1 KQ980322 KYN16561.1 KQ971321 EFA00701.2 GEZM01066536 JAV67820.1 KRK06909.1 KQ981236 KYN44316.1 KRK06906.1 KPU77468.1 KPU77467.1 KQS30028.1 KQS30021.1 GL441834 EFN64205.1 KRF94205.1 KRF94207.1 KRK06895.1 KRK06907.1 GFTR01006797 JAW09629.1 KRK06901.1 KRK06893.1 KPU77465.1 KPU77470.1 KQS30025.1 KQS30019.1 KQS30024.1 KQS30022.1 KPU77469.1 KPU77473.1 KRK06899.1 KRK06903.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007062
UP000008820
+ More
UP000007801 UP000007798 UP000192221 UP000001292 UP000008711 UP000002282 UP000000803 UP000001070 UP000009192 UP000008792 UP000075886 UP000015103 UP000095301 UP000192223 UP000007755 UP000095300 UP000053097 UP000078540 UP000279307 UP000075809 UP000078492 UP000007266 UP000078541 UP000000311
UP000007801 UP000007798 UP000192221 UP000001292 UP000008711 UP000002282 UP000000803 UP000001070 UP000009192 UP000008792 UP000075886 UP000015103 UP000095301 UP000192223 UP000007755 UP000095300 UP000053097 UP000078540 UP000279307 UP000075809 UP000078492 UP000007266 UP000078541 UP000000311
Interpro
SUPFAM
SSF54695
SSF54695
Gene 3D
ProteinModelPortal
H9J2W4
E9LP24
A0A194PLL7
A0A194QL53
A0A2A4JHA3
A0A2M4ASP2
+ More
A0A2H1V1X8 A0A1S4EVA2 A0A348G660 A0A2M4ALN5 Q7QF14 Q17PT9 A0A0P8ZSJ5 B4NPI1 A0A1W4UMX6 B4I6L8 B3NWZ6 B4Q2M4 P08510 B4JJ88 A0A1W4UNF5 B4L2A7 B4M6S3 A0A182QK67 A0A0R1EDM7 A0A0Q9W6W6 T1HFA1 P08510-3 A0A1W4V0Q2 A0A0Q9W709 A0A0Q9X091 A0A0Q5T428 A0A0R1EIB4 A0A1W4UNG0 A0A0Q9WI52 A0A1I8MXT4 A0A1W4V1X9 A0A2S1ZNM0 A0A1W4V0Q7 A0A0Q9WEM3 A0A0Q9WYW2 A0A0Q9WNB8 A0A0Q9WPL7 A0A1W4WTF4 A0A1W4UMY0 A0A1W4V0P7 W8AV96 A0A1W4WUD3 A0A0Q9W771 A0A0Q9WN85 A0A1W4X4N1 F4WVX1 A0A1I8Q8D2 A0A026WWX1 F5HJN5 P08510-9 A0A1W4X4K0 A0A1W4X443 A0A1W4V0D3 A0A0Q9WVC7 A0A1W4X2C8 W8AJ67 A0A1I8MXT7 A0A151I5G4 A0A3L8D818 A0A151WRY2 A0A1I8Q8G9 A0A1I8Q8H1 A0A0S0X7Z4 A0A1I8Q8D6 A0A1W4UMY5 A0A195DUN5 D6WHV5 P08510-7 A0A1Y1L627 A0A0R1EC30 A0A151K1K0 A0A1W4V0D8 A0A1W4UNG5 A0A0R1EIL7 A0A0N8P0L6 A0A0P8Y7Q9 A0A0Q5T3U2 A0A0Q5TEG4 E2AQU3 A0A0Q9WNA1 A0A0R1EBV3 P08510-2 A0A224XNR1 A0A0R1ECH3 A0A0R1EC16 A0A0P9C1E9 A0A0P9AI13 A0A0Q5T3T6 A0A0Q5T3R9 A0A0Q5T6N6 A0A0Q5TJD7 A0A0P8XS21 A0A0R1EFN3 A0A0R1EJQ1
A0A2H1V1X8 A0A1S4EVA2 A0A348G660 A0A2M4ALN5 Q7QF14 Q17PT9 A0A0P8ZSJ5 B4NPI1 A0A1W4UMX6 B4I6L8 B3NWZ6 B4Q2M4 P08510 B4JJ88 A0A1W4UNF5 B4L2A7 B4M6S3 A0A182QK67 A0A0R1EDM7 A0A0Q9W6W6 T1HFA1 P08510-3 A0A1W4V0Q2 A0A0Q9W709 A0A0Q9X091 A0A0Q5T428 A0A0R1EIB4 A0A1W4UNG0 A0A0Q9WI52 A0A1I8MXT4 A0A1W4V1X9 A0A2S1ZNM0 A0A1W4V0Q7 A0A0Q9WEM3 A0A0Q9WYW2 A0A0Q9WNB8 A0A0Q9WPL7 A0A1W4WTF4 A0A1W4UMY0 A0A1W4V0P7 W8AV96 A0A1W4WUD3 A0A0Q9W771 A0A0Q9WN85 A0A1W4X4N1 F4WVX1 A0A1I8Q8D2 A0A026WWX1 F5HJN5 P08510-9 A0A1W4X4K0 A0A1W4X443 A0A1W4V0D3 A0A0Q9WVC7 A0A1W4X2C8 W8AJ67 A0A1I8MXT7 A0A151I5G4 A0A3L8D818 A0A151WRY2 A0A1I8Q8G9 A0A1I8Q8H1 A0A0S0X7Z4 A0A1I8Q8D6 A0A1W4UMY5 A0A195DUN5 D6WHV5 P08510-7 A0A1Y1L627 A0A0R1EC30 A0A151K1K0 A0A1W4V0D8 A0A1W4UNG5 A0A0R1EIL7 A0A0N8P0L6 A0A0P8Y7Q9 A0A0Q5T3U2 A0A0Q5TEG4 E2AQU3 A0A0Q9WNA1 A0A0R1EBV3 P08510-2 A0A224XNR1 A0A0R1ECH3 A0A0R1EC16 A0A0P9C1E9 A0A0P9AI13 A0A0Q5T3T6 A0A0Q5T3R9 A0A0Q5T6N6 A0A0Q5TJD7 A0A0P8XS21 A0A0R1EFN3 A0A0R1EJQ1
PDB
2A79
E-value=6.05135e-135,
Score=1233
Ontologies
GO
GO:0051260
GO:0008076
GO:0005249
GO:0034765
GO:0030431
GO:0048675
GO:0045838
GO:0007637
GO:0008345
GO:0048047
GO:0060025
GO:0001508
GO:0045938
GO:0009584
GO:1903351
GO:0022843
GO:0045187
GO:0007611
GO:0006813
GO:0048150
GO:0016021
GO:0050909
GO:0007619
GO:0071805
GO:0005251
GO:0005267
GO:0005244
GO:0005216
GO:0006811
GO:0016020
GO:0030246
GO:0007169
GO:0055085
PANTHER
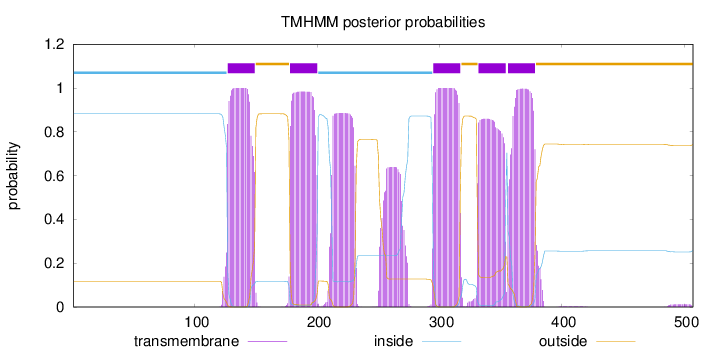
Topology
Subcellular location
Cell membrane
Length:
507
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
138.33651
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.88218
inside
1 - 126
TMhelix
127 - 149
outside
150 - 177
TMhelix
178 - 200
inside
201 - 294
TMhelix
295 - 317
outside
318 - 331
TMhelix
332 - 354
inside
355 - 355
TMhelix
356 - 378
outside
379 - 507
Population Genetic Test Statistics
Pi
18.820489
Theta
17.868116
Tajima's D
0.224611
CLR
0.045231
CSRT
0.443777811109445
Interpretation
Uncertain
