Pre Gene Modal
BGIBMGA012223
Annotation
PREDICTED:_paraplegin_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.205
Sequence
CDS
ATGTTATCAGTGAAAAGGTTCCCGGCACTTTCAGCGTCAAAATTTCAAAATAAACTACATTTTAATCTGAGTAATTATGTCTTAGGAAAGCAAGTAAACAAACATTGTGAAAGTAAGATGTTGAAATTATGTAGATCGTTCTCTGTTTTACACAATTTGACCGCCAAATCTAAGAATCCGCAACTCAGAACCTTCAATGCAGAATATAAAGCTGCTTGTGCATTACTTAAGAGGTCACAACTTCCAAGCTTGGACTCTTTCATCCACAGCATAAGCACAAGGGGCATCCACACCACACAAACCAACCATAACCAACAAGATGATAAAGATAAGGATAAAAAAGACAAGAAGAATAATGAACTCTCTTCTCTATTAATGAAAGCAGTTTTTTGGATGCTCACAACTTGTATTTTAATTATCATGAGTACTCTGCTTGTACCGAGTGACACTCCACAGAATGAGGTAATACGATATGTGTCCTGGAATGAATTTGTTTACTCGATGCTGGCGAAAGGTGAAGTTCAAGAGTTGATAGTACGACCCGATATGGAAGTTGTCACAATTATACTTTATGAGGGCGCCATCATCAAAGGAAGGCGGGCAAACAACAGAGTCTTTCACATAAACGTTGGCGACATACACAGATTCGAAGAAAAGCTACGGGAAACCGAGAAGAACCTAGGAGTCAAAGAAGGTAAAATATGTCGTGGGGTGAAATGA
Protein
MLSVKRFPALSASKFQNKLHFNLSNYVLGKQVNKHCESKMLKLCRSFSVLHNLTAKSKNPQLRTFNAEYKAACALLKRSQLPSLDSFIHSISTRGIHTTQTNHNQQDDKDKDKKDKKNNELSSLLMKAVFWMLTTCILIIMSTLLVPSDTPQNEVIRYVSWNEFVYSMLAKGEVQELIVRPDMEVVTIILYEGAIIKGRRANNRVFHINVGDIHRFEEKLRETEKNLGVKEGKICRGVK
Summary
Uniprot
A0A2W1BRD9
A0A2A4JT79
A0A212FKL2
A0A194QYA7
A0A194PFG1
V5I8W7
+ More
A0A2J7PL62 A0A2J7PL59 D6WIB7 A0A1B6KDF9 A0A1Y1L1Z2 A0A067QZP8 A0A1B6IRX5 A0A1B6CC58 A0A1B6F8D8 A0A1S4G065 Q16I01 A0A224XFM3 A0A0A9Z798 A0A0N8AGR9 A0A0P5GZB3 A0A0N8CV44 A0A0P6A453 A0A162PSJ7 A0A0N8B9E5 A0A0P5SQ79 A0A0P6HKJ4 A0A0P4VSS4 N6TDK5 A0A182GCH7 W8CD02 G3MSV1 T1I0I0 A0A224YPR2 A0A023FMC2 W8BWY1 A0A131XIU0 A0A023GP31 A0A023G039 A0A131Z542 A0A034WSE7 U4TZ15 U4TW09 A0A1E1X984 L7M7J7 A0A1I8MM13 A0A1L8D8S4 A0A0K8U9C8 A0A0K8V030 A0A0K8VBW3 A0A182LX52 A0A182W4S2 A0A1Z5L301 B0X3B3 A0A182YK71 A0A293MDT0 A0A182TB73
A0A2J7PL62 A0A2J7PL59 D6WIB7 A0A1B6KDF9 A0A1Y1L1Z2 A0A067QZP8 A0A1B6IRX5 A0A1B6CC58 A0A1B6F8D8 A0A1S4G065 Q16I01 A0A224XFM3 A0A0A9Z798 A0A0N8AGR9 A0A0P5GZB3 A0A0N8CV44 A0A0P6A453 A0A162PSJ7 A0A0N8B9E5 A0A0P5SQ79 A0A0P6HKJ4 A0A0P4VSS4 N6TDK5 A0A182GCH7 W8CD02 G3MSV1 T1I0I0 A0A224YPR2 A0A023FMC2 W8BWY1 A0A131XIU0 A0A023GP31 A0A023G039 A0A131Z542 A0A034WSE7 U4TZ15 U4TW09 A0A1E1X984 L7M7J7 A0A1I8MM13 A0A1L8D8S4 A0A0K8U9C8 A0A0K8V030 A0A0K8VBW3 A0A182LX52 A0A182W4S2 A0A1Z5L301 B0X3B3 A0A182YK71 A0A293MDT0 A0A182TB73
Pubmed
EMBL
KZ149925
PZC77608.1
NWSH01000649
PCG75029.1
AGBW02008029
OWR54278.1
+ More
KQ460953 KPJ10284.1 KQ459605 KPI92022.1 GALX01003965 JAB64501.1 NEVH01024428 PNF17074.1 PNF17073.1 KQ971321 EFA00737.1 GEBQ01030743 JAT09234.1 GEZM01066981 JAV67604.1 KK852811 KDR15916.1 GECU01018076 JAS89630.1 GEDC01026393 JAS10905.1 GECZ01023271 JAS46498.1 CH478124 EAT33893.1 GFTR01007828 JAW08598.1 GBHO01004371 GBRD01017640 JAG39233.1 JAG48187.1 GDIP01148985 JAJ74417.1 GDIQ01235592 JAK16133.1 GDIP01095895 JAM07820.1 GDIP01034894 JAM68821.1 LRGB01000446 KZS19113.1 GDIQ01199899 JAK51826.1 GDIQ01100990 JAL50736.1 GDIQ01017729 JAN77008.1 GDKW01002565 JAI54030.1 APGK01041927 APGK01041928 KB740998 ENN75828.1 JXUM01054167 KQ561808 KXJ77472.1 GAMC01002801 JAC03755.1 JO844952 AEO36569.1 ACPB03008738 GFPF01005097 MAA16243.1 GBBK01002098 JAC22384.1 GAMC01002803 JAC03753.1 GEFH01002144 JAP66437.1 GBBM01000513 JAC34905.1 GBBL01000937 JAC26383.1 GEDV01002028 JAP86529.1 GAKP01001378 JAC57574.1 KB630347 ERL83551.1 KB631202 ERL84158.1 GFAC01003368 JAT95820.1 GACK01005182 JAA59852.1 GFDF01011215 JAV02869.1 GDHF01029354 JAI22960.1 GDHF01020052 JAI32262.1 GDHF01015992 JAI36322.1 AXCM01005886 GFJQ02005241 JAW01729.1 DS232312 EDS39805.1 GFWV01019149 MAA43877.1
KQ460953 KPJ10284.1 KQ459605 KPI92022.1 GALX01003965 JAB64501.1 NEVH01024428 PNF17074.1 PNF17073.1 KQ971321 EFA00737.1 GEBQ01030743 JAT09234.1 GEZM01066981 JAV67604.1 KK852811 KDR15916.1 GECU01018076 JAS89630.1 GEDC01026393 JAS10905.1 GECZ01023271 JAS46498.1 CH478124 EAT33893.1 GFTR01007828 JAW08598.1 GBHO01004371 GBRD01017640 JAG39233.1 JAG48187.1 GDIP01148985 JAJ74417.1 GDIQ01235592 JAK16133.1 GDIP01095895 JAM07820.1 GDIP01034894 JAM68821.1 LRGB01000446 KZS19113.1 GDIQ01199899 JAK51826.1 GDIQ01100990 JAL50736.1 GDIQ01017729 JAN77008.1 GDKW01002565 JAI54030.1 APGK01041927 APGK01041928 KB740998 ENN75828.1 JXUM01054167 KQ561808 KXJ77472.1 GAMC01002801 JAC03755.1 JO844952 AEO36569.1 ACPB03008738 GFPF01005097 MAA16243.1 GBBK01002098 JAC22384.1 GAMC01002803 JAC03753.1 GEFH01002144 JAP66437.1 GBBM01000513 JAC34905.1 GBBL01000937 JAC26383.1 GEDV01002028 JAP86529.1 GAKP01001378 JAC57574.1 KB630347 ERL83551.1 KB631202 ERL84158.1 GFAC01003368 JAT95820.1 GACK01005182 JAA59852.1 GFDF01011215 JAV02869.1 GDHF01029354 JAI22960.1 GDHF01020052 JAI32262.1 GDHF01015992 JAI36322.1 AXCM01005886 GFJQ02005241 JAW01729.1 DS232312 EDS39805.1 GFWV01019149 MAA43877.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BRD9
A0A2A4JT79
A0A212FKL2
A0A194QYA7
A0A194PFG1
V5I8W7
+ More
A0A2J7PL62 A0A2J7PL59 D6WIB7 A0A1B6KDF9 A0A1Y1L1Z2 A0A067QZP8 A0A1B6IRX5 A0A1B6CC58 A0A1B6F8D8 A0A1S4G065 Q16I01 A0A224XFM3 A0A0A9Z798 A0A0N8AGR9 A0A0P5GZB3 A0A0N8CV44 A0A0P6A453 A0A162PSJ7 A0A0N8B9E5 A0A0P5SQ79 A0A0P6HKJ4 A0A0P4VSS4 N6TDK5 A0A182GCH7 W8CD02 G3MSV1 T1I0I0 A0A224YPR2 A0A023FMC2 W8BWY1 A0A131XIU0 A0A023GP31 A0A023G039 A0A131Z542 A0A034WSE7 U4TZ15 U4TW09 A0A1E1X984 L7M7J7 A0A1I8MM13 A0A1L8D8S4 A0A0K8U9C8 A0A0K8V030 A0A0K8VBW3 A0A182LX52 A0A182W4S2 A0A1Z5L301 B0X3B3 A0A182YK71 A0A293MDT0 A0A182TB73
A0A2J7PL62 A0A2J7PL59 D6WIB7 A0A1B6KDF9 A0A1Y1L1Z2 A0A067QZP8 A0A1B6IRX5 A0A1B6CC58 A0A1B6F8D8 A0A1S4G065 Q16I01 A0A224XFM3 A0A0A9Z798 A0A0N8AGR9 A0A0P5GZB3 A0A0N8CV44 A0A0P6A453 A0A162PSJ7 A0A0N8B9E5 A0A0P5SQ79 A0A0P6HKJ4 A0A0P4VSS4 N6TDK5 A0A182GCH7 W8CD02 G3MSV1 T1I0I0 A0A224YPR2 A0A023FMC2 W8BWY1 A0A131XIU0 A0A023GP31 A0A023G039 A0A131Z542 A0A034WSE7 U4TZ15 U4TW09 A0A1E1X984 L7M7J7 A0A1I8MM13 A0A1L8D8S4 A0A0K8U9C8 A0A0K8V030 A0A0K8VBW3 A0A182LX52 A0A182W4S2 A0A1Z5L301 B0X3B3 A0A182YK71 A0A293MDT0 A0A182TB73
Ontologies
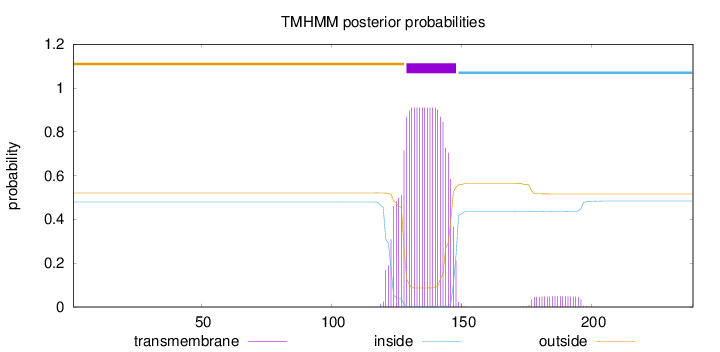
Topology
Length:
239
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.45955
Exp number, first 60 AAs:
0.00036
Total prob of N-in:
0.47900
outside
1 - 128
TMhelix
129 - 148
inside
149 - 239
Population Genetic Test Statistics
Pi
23.941563
Theta
27.353547
Tajima's D
-1.196812
CLR
0.040685
CSRT
0.102544872756362
Interpretation
Uncertain
