Pre Gene Modal
BGIBMGA012223
Annotation
PREDICTED:_paraplegin_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.104 Mitochondrial Reliability : 1.577
Sequence
CDS
ATGAATATAAATCTCGGTGGATTCAGCCAACTGGGCAGAGCTAAGTTCACTCTAGTGGATTCTATGACCGGTCAAGGCAAAGGTGTGAAATTTGAAGACGTCGCCGGTCTTAGGGAAGCCAAAATTGAAGTTATGGAATTCGTGGACTATCTAAAGAGGCCGGAGCATTACAAGAATTTGGGTGCAAAGGTACCAAAAGGTGCGCTTTTGCTTGGCCCGCCTGGCTGCGGGAAAACGTTGCTGGCTAAGGCGGTGGCTACAGAGGCCAACGTTCCGTTCCTGTCTATGAACGGTTCAGAGTTCATCGAGATGATCGGAGGCTTGGGGGCCGCTAGAGTCAGAGATCTGTTCAAAGAGGCAAGTTCTCGTAAGCCCTGCATCATATACATAGACGAGATGGACGCGGTGGGCAGAGCCAGGGCTTCCGGCACGTCCCCCTGGGGTCCAGGCGGAGGCGAATCGGAGCAGACCTTGAACCAGCTGCTTGTGGAAATGGACGGCATGAAGAGTAGGGAAGGAGTCGTTGTGTTGGCTTCGACTAACAGAGCTGATGTGCTCGATAAGGCGCTCCTCAGACCTGGCAGATTCGATCGTCACATATTAATCGACCTGCCCACCCTCCAAGAGCGCGAGGAAATTTTCGAACGACATCTAAAAAGCATTGTACTAGAAGAGCTGCCAACGTATTACATAAAGAGATTGGCTTATCTTACGCCCGGATTTAGTGGTGCAGATATAGCTAATGTGTGTAATGAGGCCGCCTTGCATGCCGCAAGATTCAAACAGAACATTGTGAAGGCAGCTAATTTAGAATACGCCGTTGAAAGAGTTGTTGGTGGTACCGAGAAACGTAGTCACGCCATGTCGCCGGCCGAGAAGAGGATAGTCGCCTACCACGAGTCCGGTCACGCACTCGTCGGCTGGCTGTTGGAACACACGGACGCTCTATTGAAAGTTACAATAGTGCCTAGAACGAACATGGCTCTAGGTTTCGCTCAGTACACGACGTCGGACCAGAAACTCTACACTAAAGAAGAGCTATTCGATCGTATGTGCATGGCGTTGGGCGGCCGTGCATCTGAAGCCATAACCTTCAATTCAATAACAAGCGGTGCGCAGAACGACTTGGAGAAAGTCACGAAAATAGCCTACGCTCAGGTGAGAGTCTATGGGATGTCGCCAACGGTTGGTCTCGTCTCCTTTCCCGATATCAACGAACGTGGCAAGAGTCCGTTCAGCAAGACCCTCAAGAACCTGATAGATCTGGAAGCCAGGAAGATAATAGCGAACGCTTATTACAGGACCGAAGATATATTGAGAACGAATCAAGATAAACTGAAGAAGCTCGCTGAAGAGCTTCTCAAGAAGGAAACATTAAACTTCAACGAAGTTCAAGGTATATTAGGTCCACCGCCCTTCCCCGGGAAGAAGTTCATCGACCCCGTAGAATTCGAGAACAGCCTCAAGAATATGGAGAAGGCGGCATCCTCTGAAGAGGGCACGCCTGTCGGAGCGCCATCAGCGAAACCAGAGTCGCAGCCTGGGTTGTGA
Protein
MNINLGGFSQLGRAKFTLVDSMTGQGKGVKFEDVAGLREAKIEVMEFVDYLKRPEHYKNLGAKVPKGALLLGPPGCGKTLLAKAVATEANVPFLSMNGSEFIEMIGGLGAARVRDLFKEASSRKPCIIYIDEMDAVGRARASGTSPWGPGGGESEQTLNQLLVEMDGMKSREGVVVLASTNRADVLDKALLRPGRFDRHILIDLPTLQEREEIFERHLKSIVLEELPTYYIKRLAYLTPGFSGADIANVCNEAALHAARFKQNIVKAANLEYAVERVVGGTEKRSHAMSPAEKRIVAYHESGHALVGWLLEHTDALLKVTIVPRTNMALGFAQYTTSDQKLYTKEELFDRMCMALGGRASEAITFNSITSGAQNDLEKVTKIAYAQVRVYGMSPTVGLVSFPDINERGKSPFSKTLKNLIDLEARKIIANAYYRTEDILRTNQDKLKKLAEELLKKETLNFNEVQGILGPPPFPGKKFIDPVEFENSLKNMEKAASSEEGTPVGAPSAKPESQPGL
Summary
Uniprot
A0A212FKL2
A0A194QYA7
A0A2A4JT79
A0A194PFG1
A0A2W1BRD9
H9JRR3
+ More
D6WIB7 A0A2J7PL62 A0A2J7PL59 A0A067QZP8 A0A1B6CC58 A0A1B6DUN7 A0A1Y1L1Z2 A0A1B6KFJ2 U4TZ15 A0A1B6F8D8 A0A0P5LDV2 A0A0N8AGR9 A0A162PSJ7 A0A0P6AXI0 A0A0P5KZH8 A0A0P6HKJ4 A0A0P5SQ79 A0A182W4S2 A0A182LX52 A0A182YK71 A0A182RV32 U4TW09 N6TDK5 A0A2P8YWV7 A0A182K6E2 A0A0P5GZB3 A0A182QMD3 A0A182J2R9 A0A1Q3FGN8 A0A1Q3FGI5 A0A182P7N0 A0A182NVI8 A0A1J1HUG4 A0A182XLR6 A0A182IA75 A0A1I8MM13 T1PPR9 A0A182VE92 A0A182LGZ0 W5JKN9 Q7QFM1 A0A182GCH7 A0A1S4G065 Q16I01 A0A336K415 A0A0N8D5Q6 B4M814 B4JKJ7 A0A232EFL9 U5EWW0 B3MRY3 O76867 K7J6T4 B4Q0X5 A0A0A1XQE0 Q9W4W8 A0A084VFN9 B3NTV6 A0A0A1X8I0 Q16V22 A0A0M5J6G5 A0A0P6A453 A0A0N8B9E5 W8BWY1 A0A0N8CV44 A0A1I8PDJ0 A0A0P5RIQ1 A0A1B6CTJ8 A0A1A9WXZ6 A0A1W4VQX8 A0A2S2NA86 B4MSH2 A0A0K8U9C8 A0A1L8D8S4 B4L634 A0A226F2K5 A0A034WSE7 A0A1L8D8U2 A0A0K8VBW3 A0A0P4VSS4 A0A069DWN2 B4H4F7 A0A1B0CZ87 Q29I07 T1I0I0 A0A0N7ZDD3 A0A224XFM3 A0A3B0K665 A0A1B0GBC2 A0A1A9YH90 A0A1A9VP34 E2C000 A0A0V0G7H8 A0A2M4BEJ3
D6WIB7 A0A2J7PL62 A0A2J7PL59 A0A067QZP8 A0A1B6CC58 A0A1B6DUN7 A0A1Y1L1Z2 A0A1B6KFJ2 U4TZ15 A0A1B6F8D8 A0A0P5LDV2 A0A0N8AGR9 A0A162PSJ7 A0A0P6AXI0 A0A0P5KZH8 A0A0P6HKJ4 A0A0P5SQ79 A0A182W4S2 A0A182LX52 A0A182YK71 A0A182RV32 U4TW09 N6TDK5 A0A2P8YWV7 A0A182K6E2 A0A0P5GZB3 A0A182QMD3 A0A182J2R9 A0A1Q3FGN8 A0A1Q3FGI5 A0A182P7N0 A0A182NVI8 A0A1J1HUG4 A0A182XLR6 A0A182IA75 A0A1I8MM13 T1PPR9 A0A182VE92 A0A182LGZ0 W5JKN9 Q7QFM1 A0A182GCH7 A0A1S4G065 Q16I01 A0A336K415 A0A0N8D5Q6 B4M814 B4JKJ7 A0A232EFL9 U5EWW0 B3MRY3 O76867 K7J6T4 B4Q0X5 A0A0A1XQE0 Q9W4W8 A0A084VFN9 B3NTV6 A0A0A1X8I0 Q16V22 A0A0M5J6G5 A0A0P6A453 A0A0N8B9E5 W8BWY1 A0A0N8CV44 A0A1I8PDJ0 A0A0P5RIQ1 A0A1B6CTJ8 A0A1A9WXZ6 A0A1W4VQX8 A0A2S2NA86 B4MSH2 A0A0K8U9C8 A0A1L8D8S4 B4L634 A0A226F2K5 A0A034WSE7 A0A1L8D8U2 A0A0K8VBW3 A0A0P4VSS4 A0A069DWN2 B4H4F7 A0A1B0CZ87 Q29I07 T1I0I0 A0A0N7ZDD3 A0A224XFM3 A0A3B0K665 A0A1B0GBC2 A0A1A9YH90 A0A1A9VP34 E2C000 A0A0V0G7H8 A0A2M4BEJ3
Pubmed
22118469
26354079
28756777
19121390
18362917
19820115
+ More
24845553 28004739 23537049 25244985 29403074 25315136 20966253 20920257 23761445 12364791 14747013 17210077 26483478 17510324 17994087 28648823 20075255 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 24495485 25348373 27129103 26334808 15632085 20798317
24845553 28004739 23537049 25244985 29403074 25315136 20966253 20920257 23761445 12364791 14747013 17210077 26483478 17510324 17994087 28648823 20075255 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 24495485 25348373 27129103 26334808 15632085 20798317
EMBL
AGBW02008029
OWR54278.1
KQ460953
KPJ10284.1
NWSH01000649
PCG75029.1
+ More
KQ459605 KPI92022.1 KZ149925 PZC77608.1 BABH01017326 BABH01017327 BABH01017328 BABH01017329 KQ971321 EFA00737.1 NEVH01024428 PNF17074.1 PNF17073.1 KK852811 KDR15916.1 GEDC01026393 JAS10905.1 GEDC01007902 JAS29396.1 GEZM01066981 JAV67604.1 GEBQ01030023 JAT09954.1 KB630347 ERL83551.1 GECZ01023271 JAS46498.1 GDIQ01171002 JAK80723.1 GDIP01148985 JAJ74417.1 LRGB01000446 KZS19113.1 GDIP01023308 JAM80407.1 GDIQ01185100 JAK66625.1 GDIQ01017729 JAN77008.1 GDIQ01100990 JAL50736.1 AXCM01005886 KB631202 ERL84158.1 APGK01041927 APGK01041928 KB740998 ENN75828.1 PYGN01000311 PSN48736.1 GDIQ01235592 JAK16133.1 AXCN02002225 GFDL01008334 JAV26711.1 GFDL01008386 JAV26659.1 CVRI01000017 CRK90142.1 APCN01001131 KA650195 AFP64824.1 ADMH02001263 ETN63344.1 AAAB01008846 EAA06300.5 JXUM01054167 KQ561808 KXJ77472.1 CH478124 EAT33893.1 UFQS01000107 UFQT01000107 SSW99714.1 SSX20094.1 GDIP01066199 JAM37516.1 CH940653 EDW62290.2 CH916370 EDW00100.1 NNAY01004963 OXU17169.1 GANO01002792 JAB57079.1 CH902622 EDV34538.1 AL023874 CAA19646.1 AAZX01008957 CM000162 EDX01342.1 GBXI01001544 JAD12748.1 AE014298 AAF45806.1 AAS65257.2 ATLV01012451 KE524793 KFB36783.1 CH954180 EDV45664.1 GBXI01007091 JAD07201.1 CH477606 EAT38370.1 CP012528 ALC49898.1 GDIP01034894 JAM68821.1 GDIQ01199899 JAK51826.1 GAMC01002803 JAC03753.1 GDIP01095895 JAM07820.1 GDIQ01100234 JAL51492.1 GEDC01020597 JAS16701.1 GGMR01001436 MBY14055.1 CH963851 EDW75061.1 GDHF01029354 JAI22960.1 GFDF01011215 JAV02869.1 CH933812 EDW05830.1 LNIX01000001 OXA63570.1 GAKP01001378 JAC57574.1 GFDF01011216 JAV02868.1 GDHF01015992 JAI36322.1 GDKW01002565 JAI54030.1 GBGD01000643 JAC88246.1 CH479209 EDW32708.1 AJVK01002128 CH379064 EAL31599.2 ACPB03008738 GDRN01044354 GDRN01044353 JAI66928.1 GFTR01007828 JAW08598.1 OUUW01000025 SPP89625.1 CCAG010022538 GL451712 EFN78696.1 GECL01002203 JAP03921.1 GGFJ01002324 MBW51465.1
KQ459605 KPI92022.1 KZ149925 PZC77608.1 BABH01017326 BABH01017327 BABH01017328 BABH01017329 KQ971321 EFA00737.1 NEVH01024428 PNF17074.1 PNF17073.1 KK852811 KDR15916.1 GEDC01026393 JAS10905.1 GEDC01007902 JAS29396.1 GEZM01066981 JAV67604.1 GEBQ01030023 JAT09954.1 KB630347 ERL83551.1 GECZ01023271 JAS46498.1 GDIQ01171002 JAK80723.1 GDIP01148985 JAJ74417.1 LRGB01000446 KZS19113.1 GDIP01023308 JAM80407.1 GDIQ01185100 JAK66625.1 GDIQ01017729 JAN77008.1 GDIQ01100990 JAL50736.1 AXCM01005886 KB631202 ERL84158.1 APGK01041927 APGK01041928 KB740998 ENN75828.1 PYGN01000311 PSN48736.1 GDIQ01235592 JAK16133.1 AXCN02002225 GFDL01008334 JAV26711.1 GFDL01008386 JAV26659.1 CVRI01000017 CRK90142.1 APCN01001131 KA650195 AFP64824.1 ADMH02001263 ETN63344.1 AAAB01008846 EAA06300.5 JXUM01054167 KQ561808 KXJ77472.1 CH478124 EAT33893.1 UFQS01000107 UFQT01000107 SSW99714.1 SSX20094.1 GDIP01066199 JAM37516.1 CH940653 EDW62290.2 CH916370 EDW00100.1 NNAY01004963 OXU17169.1 GANO01002792 JAB57079.1 CH902622 EDV34538.1 AL023874 CAA19646.1 AAZX01008957 CM000162 EDX01342.1 GBXI01001544 JAD12748.1 AE014298 AAF45806.1 AAS65257.2 ATLV01012451 KE524793 KFB36783.1 CH954180 EDV45664.1 GBXI01007091 JAD07201.1 CH477606 EAT38370.1 CP012528 ALC49898.1 GDIP01034894 JAM68821.1 GDIQ01199899 JAK51826.1 GAMC01002803 JAC03753.1 GDIP01095895 JAM07820.1 GDIQ01100234 JAL51492.1 GEDC01020597 JAS16701.1 GGMR01001436 MBY14055.1 CH963851 EDW75061.1 GDHF01029354 JAI22960.1 GFDF01011215 JAV02869.1 CH933812 EDW05830.1 LNIX01000001 OXA63570.1 GAKP01001378 JAC57574.1 GFDF01011216 JAV02868.1 GDHF01015992 JAI36322.1 GDKW01002565 JAI54030.1 GBGD01000643 JAC88246.1 CH479209 EDW32708.1 AJVK01002128 CH379064 EAL31599.2 ACPB03008738 GDRN01044354 GDRN01044353 JAI66928.1 GFTR01007828 JAW08598.1 OUUW01000025 SPP89625.1 CCAG010022538 GL451712 EFN78696.1 GECL01002203 JAP03921.1 GGFJ01002324 MBW51465.1
Proteomes
UP000007151
UP000053240
UP000218220
UP000053268
UP000005204
UP000007266
+ More
UP000235965 UP000027135 UP000030742 UP000076858 UP000075920 UP000075883 UP000076408 UP000075900 UP000019118 UP000245037 UP000075881 UP000075886 UP000075880 UP000075885 UP000075884 UP000183832 UP000076407 UP000075840 UP000095301 UP000075903 UP000075882 UP000000673 UP000007062 UP000069940 UP000249989 UP000008820 UP000008792 UP000001070 UP000215335 UP000007801 UP000002358 UP000002282 UP000000803 UP000030765 UP000008711 UP000092553 UP000095300 UP000091820 UP000192221 UP000007798 UP000009192 UP000198287 UP000008744 UP000092462 UP000001819 UP000015103 UP000268350 UP000092444 UP000092443 UP000078200 UP000008237
UP000235965 UP000027135 UP000030742 UP000076858 UP000075920 UP000075883 UP000076408 UP000075900 UP000019118 UP000245037 UP000075881 UP000075886 UP000075880 UP000075885 UP000075884 UP000183832 UP000076407 UP000075840 UP000095301 UP000075903 UP000075882 UP000000673 UP000007062 UP000069940 UP000249989 UP000008820 UP000008792 UP000001070 UP000215335 UP000007801 UP000002358 UP000002282 UP000000803 UP000030765 UP000008711 UP000092553 UP000095300 UP000091820 UP000192221 UP000007798 UP000009192 UP000198287 UP000008744 UP000092462 UP000001819 UP000015103 UP000268350 UP000092444 UP000092443 UP000078200 UP000008237
Interpro
Gene 3D
ProteinModelPortal
A0A212FKL2
A0A194QYA7
A0A2A4JT79
A0A194PFG1
A0A2W1BRD9
H9JRR3
+ More
D6WIB7 A0A2J7PL62 A0A2J7PL59 A0A067QZP8 A0A1B6CC58 A0A1B6DUN7 A0A1Y1L1Z2 A0A1B6KFJ2 U4TZ15 A0A1B6F8D8 A0A0P5LDV2 A0A0N8AGR9 A0A162PSJ7 A0A0P6AXI0 A0A0P5KZH8 A0A0P6HKJ4 A0A0P5SQ79 A0A182W4S2 A0A182LX52 A0A182YK71 A0A182RV32 U4TW09 N6TDK5 A0A2P8YWV7 A0A182K6E2 A0A0P5GZB3 A0A182QMD3 A0A182J2R9 A0A1Q3FGN8 A0A1Q3FGI5 A0A182P7N0 A0A182NVI8 A0A1J1HUG4 A0A182XLR6 A0A182IA75 A0A1I8MM13 T1PPR9 A0A182VE92 A0A182LGZ0 W5JKN9 Q7QFM1 A0A182GCH7 A0A1S4G065 Q16I01 A0A336K415 A0A0N8D5Q6 B4M814 B4JKJ7 A0A232EFL9 U5EWW0 B3MRY3 O76867 K7J6T4 B4Q0X5 A0A0A1XQE0 Q9W4W8 A0A084VFN9 B3NTV6 A0A0A1X8I0 Q16V22 A0A0M5J6G5 A0A0P6A453 A0A0N8B9E5 W8BWY1 A0A0N8CV44 A0A1I8PDJ0 A0A0P5RIQ1 A0A1B6CTJ8 A0A1A9WXZ6 A0A1W4VQX8 A0A2S2NA86 B4MSH2 A0A0K8U9C8 A0A1L8D8S4 B4L634 A0A226F2K5 A0A034WSE7 A0A1L8D8U2 A0A0K8VBW3 A0A0P4VSS4 A0A069DWN2 B4H4F7 A0A1B0CZ87 Q29I07 T1I0I0 A0A0N7ZDD3 A0A224XFM3 A0A3B0K665 A0A1B0GBC2 A0A1A9YH90 A0A1A9VP34 E2C000 A0A0V0G7H8 A0A2M4BEJ3
D6WIB7 A0A2J7PL62 A0A2J7PL59 A0A067QZP8 A0A1B6CC58 A0A1B6DUN7 A0A1Y1L1Z2 A0A1B6KFJ2 U4TZ15 A0A1B6F8D8 A0A0P5LDV2 A0A0N8AGR9 A0A162PSJ7 A0A0P6AXI0 A0A0P5KZH8 A0A0P6HKJ4 A0A0P5SQ79 A0A182W4S2 A0A182LX52 A0A182YK71 A0A182RV32 U4TW09 N6TDK5 A0A2P8YWV7 A0A182K6E2 A0A0P5GZB3 A0A182QMD3 A0A182J2R9 A0A1Q3FGN8 A0A1Q3FGI5 A0A182P7N0 A0A182NVI8 A0A1J1HUG4 A0A182XLR6 A0A182IA75 A0A1I8MM13 T1PPR9 A0A182VE92 A0A182LGZ0 W5JKN9 Q7QFM1 A0A182GCH7 A0A1S4G065 Q16I01 A0A336K415 A0A0N8D5Q6 B4M814 B4JKJ7 A0A232EFL9 U5EWW0 B3MRY3 O76867 K7J6T4 B4Q0X5 A0A0A1XQE0 Q9W4W8 A0A084VFN9 B3NTV6 A0A0A1X8I0 Q16V22 A0A0M5J6G5 A0A0P6A453 A0A0N8B9E5 W8BWY1 A0A0N8CV44 A0A1I8PDJ0 A0A0P5RIQ1 A0A1B6CTJ8 A0A1A9WXZ6 A0A1W4VQX8 A0A2S2NA86 B4MSH2 A0A0K8U9C8 A0A1L8D8S4 B4L634 A0A226F2K5 A0A034WSE7 A0A1L8D8U2 A0A0K8VBW3 A0A0P4VSS4 A0A069DWN2 B4H4F7 A0A1B0CZ87 Q29I07 T1I0I0 A0A0N7ZDD3 A0A224XFM3 A0A3B0K665 A0A1B0GBC2 A0A1A9YH90 A0A1A9VP34 E2C000 A0A0V0G7H8 A0A2M4BEJ3
PDB
6NYY
E-value=5.2474e-129,
Score=1182
Ontologies
GO
Topology
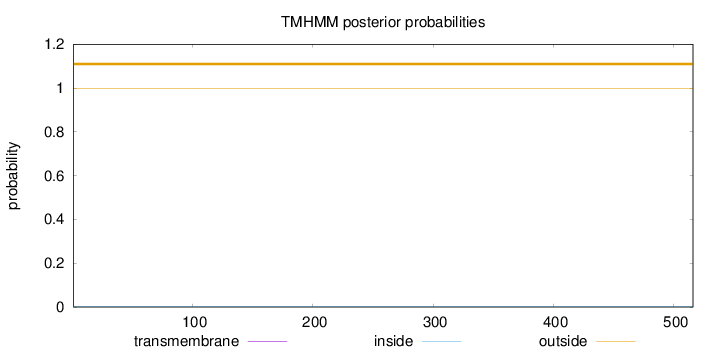
Length:
516
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00453
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00075
outside
1 - 516
Population Genetic Test Statistics
Pi
27.229046
Theta
19.62718
Tajima's D
-1.537076
CLR
0.007357
CSRT
0.055597220138993
Interpretation
Uncertain
