Gene
KWMTBOMO00601
Annotation
S33901_reverse_transcriptase_-_silkworm_transposon_Pao
Location in the cell
Nuclear Reliability : 2.408
Sequence
CDS
ATGGTGCTCACACGGCCCGAGAAGATGAGGTCGTCACGCCTTGACCGGCCCGGCAACGAAGCTGTCTCGGGCTCAACATCACAAGACGCACAGCAACGAGCTGTGTCGGCCGCGCCGCGTTCGGGAGTGACGCAGGGACCGCCACCGTTGGCGGCAACCTCCTCCCAGTCAACCGCGCCGCGTTCGGGAGTGACGCAGGGACCGCCACCGATGGCGGCACCCTCCTCCCGAGTGGAAAAGTGTCCGGGAGTGCCGCGGGGCTCACAAGCGATGCCCCCCTCCCGCCAAATATTGCCTACGCAATACTCGCCGGCGTCGCCAACGGGACCGAACAGCGGGGTCTGTCGTCGAGTGGTCAGACCTCCCAGCACCGCGACACCGGCAACAACTTCAGGCGACGCAGCAACGGAGAGCGTCGTATCCGCGAGCCAGAACAGGAAAGAGTCAGCGCTCACGGCCAATCCAATTGTACACGGGTCCCACATCACGGACGCCGCTGCCAAAATCTCGACGACAGAAGAAGGCTCCACACTCCGCGACGGGATGGCGCCACCAGTACGAGGATCTTCAAGAAAGTCGAGTCAACAGTCACAACGGCTGTTGCTTATGGCGCGCCTCAAAGAAGAGCATCTTCGCGCCAAAGAAGAACAAGCCCGACTCCAAGCAGAGCTCGCCGCCGCCCGCATCTCAACATTAGAAGCCGAAGCGCTCGTTGAAGAGGAGGAAGATGCGCGCACAACACTCACGGAGGACGAGAACGAGCAATCACAGCGCATCGACACATGGCTCAGTCAGCAACGATCGATCGCGCTACAGGGAGTCGAAGAAAGAGTAATGCAGCCAACACACGGATCACCAGCCACCATCGAGCAACCGCAACTGGAACTTCCCGCACCAATAGAACAGCTGAAGCTACCGGCCCCTGCCGCGGCACCGATTGCCGCACCTGTCGCACCGAAGAGCGACATCGCCGAACTAGCAGCTGCCATCGCGACGGCCGCCCGCCAAGCCAGGCCCGGACCATCGCATCGGTACTACGGGGAGCTTCCAGTGTACAGTGGATCGCACCAGGAATGGCTCTCCTTCAAGGTCGCCTACGCCGAATCAGCGGACAGCTTCAGCGCCGCAGAGAACACTGCGAGACTTCGGCGTACGTTAAGAGGAAGAGCAAGAGAAGCGGTCGAAAACTTGTTGCTGCATTACACCGAGCCCGCCGAGATCATGCGGACTCTAGAATCGCGCTTCGGACGTCCAGAAGCAATCGCCGCAACGGAGCTGGAACGACTGCGCGCGCTGCCACGCTGCACGGACACACCGAGGGACATCTGCGTATTCGCGAACAAGGTGAACAACGTCGTCGCCGCTCTGAGGGCCCTCGACCGCGTACATTATATGTACAACCCGGAGCTCACCAACATCACATCAGAGAAGCTGCCGTCCACTCTACGCCACCGCTGGTTCGAATTCTCGGCAACTCAACCGGCGGGAGAACCGGACCTCGTCAAGCTGTCGCGCTTCTTACAGCGCGAGGCGGACCTGTGCAGCCCATACGCACAGCCGGAACCAGAGACGAGAGTGGAGAACACCAGCGGTCGGCGGAAGATGGTGAACACGCCTCAGAGGACGCACACCACGCAAGCGAAGGAAGAGAGGAAATGCGCAGCATGTCAGAAGCCGGGGCACATCCCACAAGATTGTCCCGAATTCAAGAGAGCAGCCGTCAGCGAGCGCTGGGATACGGCGAAGCGCGAGAATTTGTGTTTCCGGTGCCTACGGTTCCGGTCTCGGGGACACGTATGCAAAAAGAAGAAGTGCGGCGTCGACAGCTGTGAACGCACGCACCACGAGCTGTTGCATAAGAAGCCAGCATGGCAGAAGCCGAAAGAAAAAGAAGAGGCGGTCACCTCGACGTGGGCCGCAAGAAGTGCGACAGCCTACCTGAAGATGGCGCCAATTACTGTTATCGGACCGGCGGGAGAAGCAGATACATGGGCCCTGCTGGACGACGGGTCGACGATCTCGCTGATTGACGAAGACTTCGCGAGACGAGTGGGCGCCAAAGGACCGATTGAACCCCTCTTCATCACTGCCATCGGAGACAATAAGATAGACGCAACACGCTCACGACGCGTCCCGCTGAAGCTATGCGGACGCACGGGGGAACCGCAAGAACTGAACCTACGGACAGTCAACGGCTTAAAATTAACACCACAGCGACTGAACATCGAAGTCCTCGCGTCGTGCCATCACCTCACCGACCTCCGGCATCACTTCAAAACGAGCTACGCCTCGCCGAAGATACTGATCGGCCAAGACAACTGGCACCTGCTGGTGACGGAGGAGATGCGGACGGGACGACGCGATCAACCAGTGGCGTCTCGTACTCCGCTCGGGTGGGTCGTGCACGGAGCACACCCGGGAGGCAAGAGACAGCGCGTCAACTTCGTGGCACATGCGACGACGGTCGACACGAACGTGGACGAAGCCCTCAAGCACTACGTCGCGATTGAGGGACTCACCGTCGCCGCAAAGACGCCGAAGAACGATCCGGACGAGCGCGCGCTGAAGATCCTGCGAGAGACCACTCAACAGCGACCCGACGGCCGCTACGAGACCGCACTGCTGTGGCGTGAGGAGAGCCTGAGAATGCCCAACAATTTTGAAGCCGCAATAAATCGCCTGACGTCCGTAGAAAAGAAACTGGAGAAGGATCCAAATTTGAAAGAGCGCTACAAGCGACAAATGGACGCACTAGTAGCGAAAGGATACGCCGAAGTAGCTCCGTCAACGGGGACGAAGGACCGAACCTGGTACCTGCCACACTTCGGCGTAACACAACCAATGAAGCAGGAGAAGCTTCGCATCGCGCACAACGCCGCTGCGAAAACTAGAGGGAAGAAGCCCGAACGACTTCCTGCTCACCGACCCGGACCTGCTGCAGTCGCTGCCCGGAGCGATGATGCACTTCAGGCAGCACGCCGTTGCCGTCTCCGCGGACATCGCGGAGATGTTCATCCGGATAGGCGTCCGAAGCGAGGACCGCGATGCGCTCCGGTACTTGTGGAGGGAGGACCCTTCACGAGAGCCCACAGAATACCGGACGAGGTCCATCGTCTTCGGCGCGACAAGGTCACCTGCAACCGCCATCTAAGTGAAGAACCGACTGCAGTACCCGGACGCCGCCGACCTCAAGCTCCACACAAAGCCAGACAGACACGAGTGCTCTGGACCGACAGCGCCACCATAGAAGAAAACAGCACTGTGGAGGAATGGCGCTGGGTCCCCACAAGAGAGAATGTCGCCGACGACGCCACAAGAGGAATACCCGTCGGCTTTGAAAGAAACCACCGCTGGTTCATAGGCCCGGAATACCTAAGACGGCATCCGGATGATTGGCTCGTACAACGGACTGCAAGAAGGGCAGAAGAGACGGGTGAAGAGAAGTGCGCAACGCTCGCCGTGAGCAGAGAGAGCCTCGGCGAAGCGATCCCGGACCCAAGACGCTTCTCCAAGTGGGAGAAGTACCTGCGTGCAACGGGGCGAATACTACAATTCGTCAGCCTATGCCGCAGGAGTCGCGAGCGCACTCATTACAAGAGGACCAGGCGGAACCCGCGTTCGGACCCGACGTGGGAGAAGAATAGAAAGAAGACGACTTCGAAGACCCCGCTGAACACACCGCGGACGCCAGAGCAAGCAGTCATCCAATGGAAGACTTTAGACGCCGACACGCTTCGACGCGCCGAGACGCTCATTTGGCGACAGAGCCAGCGGGCGGCCTTCGAGGAAGAGATTTTGACGCTCCAGCGAGGGAAACAAATATCCCCAGCGAGCCGACTGCGAAACTTGTCCGTAACTTACGCGGACGGGATATTGAAAATTAACGGACGCATCGGCAACATAGAGGGAGCTGACGTCATCACCTCGCCTCTCGTGCTAGACGGATCCCGACGCGAAACGAGACTGATGATAGACTTCATCCATAGAAAGATGCACCACGCCGGAACCGAAGCTACGATCGCCGAGTGCCGACAGTCAAGCACCCAGAAGCGCTACGTGGCCATCTTCACATGTCTCACTGCGCGTGCGGTACACTTAGAACCGGCAGCGAGCCTCAGCACGGACTCAGCGGTGATGGCACTCCGGCGCATGATCGCGCGCCGGGGAGCCCCGACGGAGATATGGAGCGACAACGGCACCAATTTACGAGGTGCCGACAAGGAGCTGCGCCAAGCTATGGACAAGGCGACTGAGCATGAAGCGAGCTTAAGACTTATCCAATGGCGCTTCATCCCACCGGGCGCGCCTTTCATGGGCGGCGCCTGGGAAAGAATGGTGCGGGCGGTAAAGGCCGCGCTCTCGGCCACGGAACAGCCGAGGCACCCGACGCCCGAAATATTTCACACTCTGCTAGCGGAGGCAGAGTTCACAGTGAACAGCCGACCTCTCACTCACGTATCGGTGAGCGCCGACGATCCCGACCCTCTAACACCGAACCACTTCCTGCTGGGAGGCCCGGCGCGAGTCCCCGTGCCGGGCAAGTTTGACGACGCCGACCTCATAGGGCGCGCACACTGGCGCGCAGCTCAACGTCTGGCAGATGTGTTCTGGAGTCGCTGGCTCCGGGAGTACCTGCCCGACCTGCAGAACCGGCGGGAGCCCCACAGCCGCGGGCCCGCCCTGAAGATAGGCGACGTCGTGATAATAGCAGACGGAACGCTCCCACGGAACACCTGGCCACGAGGGATCATCCAGGAGGTTTACCCGGGGGCCGACGGCATCACTCGAGTGGTCGACGTGCGCACCGCCGGCGGTATCCTGCGACGCCCGGCAAAGAAGATCATCGTGCTGCCCACGATGTCCGCCGCTCACCAGGGAGGATGCAGCGACGTGAACGACGCTGCACGGCGGGAGGATGTTCGCGACGGACATGAATAA
Protein
MVLTRPEKMRSSRLDRPGNEAVSGSTSQDAQQRAVSAAPRSGVTQGPPPLAATSSQSTAPRSGVTQGPPPMAAPSSRVEKCPGVPRGSQAMPPSRQILPTQYSPASPTGPNSGVCRRVVRPPSTATPATTSGDAATESVVSASQNRKESALTANPIVHGSHITDAAAKISTTEEGSTLRDGMAPPVRGSSRKSSQQSQRLLLMARLKEEHLRAKEEQARLQAELAAARISTLEAEALVEEEEDARTTLTEDENEQSQRIDTWLSQQRSIALQGVEERVMQPTHGSPATIEQPQLELPAPIEQLKLPAPAAAPIAAPVAPKSDIAELAAAIATAARQARPGPSHRYYGELPVYSGSHQEWLSFKVAYAESADSFSAAENTARLRRTLRGRAREAVENLLLHYTEPAEIMRTLESRFGRPEAIAATELERLRALPRCTDTPRDICVFANKVNNVVAALRALDRVHYMYNPELTNITSEKLPSTLRHRWFEFSATQPAGEPDLVKLSRFLQREADLCSPYAQPEPETRVENTSGRRKMVNTPQRTHTTQAKEERKCAACQKPGHIPQDCPEFKRAAVSERWDTAKRENLCFRCLRFRSRGHVCKKKKCGVDSCERTHHELLHKKPAWQKPKEKEEAVTSTWAARSATAYLKMAPITVIGPAGEADTWALLDDGSTISLIDEDFARRVGAKGPIEPLFITAIGDNKIDATRSRRVPLKLCGRTGEPQELNLRTVNGLKLTPQRLNIEVLASCHHLTDLRHHFKTSYASPKILIGQDNWHLLVTEEMRTGRRDQPVASRTPLGWVVHGAHPGGKRQRVNFVAHATTVDTNVDEALKHYVAIEGLTVAAKTPKNDPDERALKILRETTQQRPDGRYETALLWREESLRMPNNFEAAINRLTSVEKKLEKDPNLKERYKRQMDALVAKGYAEVAPSTGTKDRTWYLPHFGVTQPMKQEKLRIAHNAAAKTRGKKPERLPAHRPGPAAVAARSDDALQAARRCRLRGHRGDVHPDRRPKRGPRCAPVLVEGGPFTRAHRIPDEVHRLRRDKVTCNRHLSEEPTAVPGRRRPQAPHKARQTRVLWTDSATIEENSTVEEWRWVPTRENVADDATRGIPVGFERNHRWFIGPEYLRRHPDDWLVQRTARRAEETGEEKCATLAVSRESLGEAIPDPRRFSKWEKYLRATGRILQFVSLCRRSRERTHYKRTRRNPRSDPTWEKNRKKTTSKTPLNTPRTPEQAVIQWKTLDADTLRRAETLIWRQSQRAAFEEEILTLQRGKQISPASRLRNLSVTYADGILKINGRIGNIEGADVITSPLVLDGSRRETRLMIDFIHRKMHHAGTEATIAECRQSSTQKRYVAIFTCLTARAVHLEPAASLSTDSAVMALRRMIARRGAPTEIWSDNGTNLRGADKELRQAMDKATEHEASLRLIQWRFIPPGAPFMGGAWERMVRAVKAALSATEQPRHPTPEIFHTLLAEAEFTVNSRPLTHVSVSADDPDPLTPNHFLLGGPARVPVPGKFDDADLIGRAHWRAAQRLADVFWSRWLREYLPDLQNRREPHSRGPALKIGDVVIIADGTLPRNTWPRGIIQEVYPGADGITRVVDVRTAGGILRRPAKKIIVLPTMSAAHQGGCSDVNDAARREDVRDGHE
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
Topology
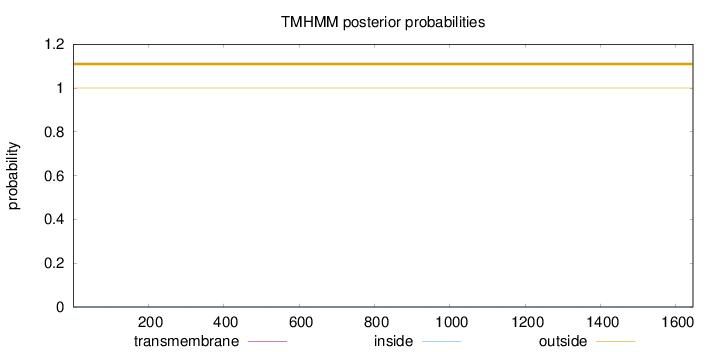
Length:
1645
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0011
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00000
outside
1 - 1645
Population Genetic Test Statistics
Pi
25.315194
Theta
28.407661
Tajima's D
-1.6073
CLR
0.000086
CSRT
0.0467976601169941
Interpretation
Uncertain
