Gene
KWMTBOMO00595
Pre Gene Modal
BGIBMGA012227
Annotation
PREDICTED:_uncharacterized_protein_LOC105842554_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.109
Sequence
CDS
ATGAGGGTTGACTTCCGCGGGCTAAAGTGTGCGACTGCACTCCGAGCCAATCTCAAATCTGCAATGTACGAAAGCAGCTGCTCCTATGCTGGGGCTTTGGAACTAAAACGCAGCGCACTGAAGGAGGAACCGTGGCCGGCATCAGAATGGCCACCCTATCGCCTTCACCAATCAACCTTCGAGCAATTGAAGCAGAGCGTGGAGAAGGCCAAGGCGGCTCTTCAGGACAGGTCAGGCTTACTCGGGACGGCGACGGCTTTCGGTGACTTCTACGGAGGTCAGCTGGGCCAAGACGCTGGAGCTACACTAGGCTTCGGACGTGGCGCTCGAACAGAAGATTCTATGCTCGCATCTGTGGACAAAGGCAAAACTGTTGGCAACGATAAACTCTCTTCGGGGCTCAGTAAGGGGTACGCCACACCCACACTCTCAGACACATTGCGCAGCACTAAATGTACAGGTCAGTGGAATTTCTCTTTTTTAAAACGTGCGTATGCATAA
Protein
MRVDFRGLKCATALRANLKSAMYESSCSYAGALELKRSALKEEPWPASEWPPYRLHQSTFEQLKQSVEKAKAALQDRSGLLGTATAFGDFYGGQLGQDAGATLGFGRGARTEDSMLASVDKGKTVGNDKLSSGLSKGYATPTLSDTLRSTKCTGQWNFSFLKRAYA
Summary
Similarity
Belongs to the ETS family.
Uniprot
EMBL
Proteomes
Pfam
PF00178 Ets
Interpro
SUPFAM
SSF46785
SSF46785
Gene 3D
ProteinModelPortal
Ontologies
PANTHER
Topology
Subcellular location
Nucleus
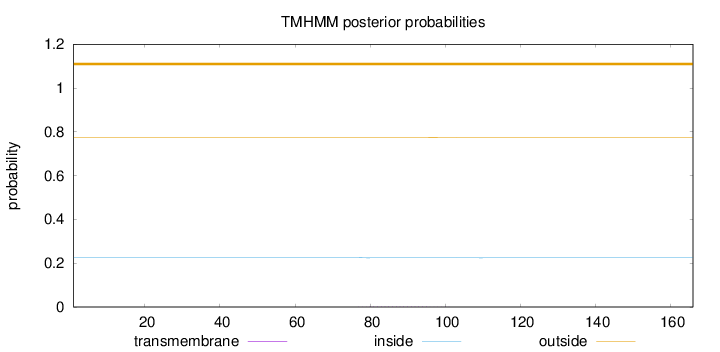
Length:
166
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02713
Exp number, first 60 AAs:
0.00104
Total prob of N-in:
0.22590
outside
1 - 166
Population Genetic Test Statistics
Pi
23.769147
Theta
22.391731
Tajima's D
-1.024393
CLR
0.004942
CSRT
0.130193490325484
Interpretation
Uncertain
