Gene
KWMTBOMO00585
Pre Gene Modal
BGIBMGA012234
Annotation
PREDICTED:_LIM_homeobox_transcription_factor_1-beta_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.221
Sequence
CDS
ATGCGGGTCGGAGAGCTCTCCTGGCATGAGCACTGCCTCAGCTGCTGTGTCTGCGGGTGTCCGCTTGCTCACACCTGCTACACCAGAAACGCGAAACTTTACTGCAAACCCGATTACGATAGATTGTTCGGGGTGAAGTGTACAAGATGCGGCGACCGTCTTCTCCCACAAGAAATGGTAATGAGGGCCCAACAGTATGTCTTTCACATACAATGCTTCGTCTGCGTGATGTGTTGCCAACCGCTACAGAAGGGCGAACAGTACGTTATTCGCGCTGGTCAAATATTTTGCAGGCAAGACTTTGAAAAAGAAATGTATCTAATGCAGCATGCTGAAGATGACATGATTATCGATGATTCAGAGCGTCCTAGAGACGGACGACGAGGCCCGAAGCGTCCTCGTACCATCCTTACTTCTGCCCAACGCAGGCAGTTCAAAGCGTCTTTCGAGGTCAGTCCGAAGCCTTGCAGGAAGGTCCGAGAGGCACTCGCTAAAGACACCGGTCTGAGTGTTAGGGTCGTGCAAGTTTGGTTTCAAAATCAACGAGCTAAAATGAAAAAGATCCAACGCAAAGCCAAACAAGAAGGCGACAAAAACGGTGACAAAGAGAAAGATAAGGACGAAAAATCGATCAAACAAGAGTCGCCGACAAGCGAGCACGGAAATTATTTAGGTCTTGATGGATCATACTCTGCCTCCAGCCAGCCATTAAATCCTAACCTTCCATATTCGCCCGATGATTACCCAGCGCATTCAGGGGACAGCTTTTGCAGTTCCGACATATCCCTCGATGGCAGCAATTTCGACCAGCTAGACGAGGGCGCGTCCGACACAATGAGCCTGCAGAATCTGGAAGTGCAGCACCACAATCACCAGCATCCTGGCCACTCTGCACACGAGCCACTGAACCTCGGAACCGGAGCCGTCGTTAACCCGATCGACAAACTTTACCTCATGCAGAATTCATATTTTAGTACTGATCATTGA
Protein
MRVGELSWHEHCLSCCVCGCPLAHTCYTRNAKLYCKPDYDRLFGVKCTRCGDRLLPQEMVMRAQQYVFHIQCFVCVMCCQPLQKGEQYVIRAGQIFCRQDFEKEMYLMQHAEDDMIIDDSERPRDGRRGPKRPRTILTSAQRRQFKASFEVSPKPCRKVREALAKDTGLSVRVVQVWFQNQRAKMKKIQRKAKQEGDKNGDKEKDKDEKSIKQESPTSEHGNYLGLDGSYSASSQPLNPNLPYSPDDYPAHSGDSFCSSDISLDGSNFDQLDEGASDTMSLQNLEVQHHNHQHPGHSAHEPLNLGTGAVVNPIDKLYLMQNSYFSTDH
Summary
Uniprot
H9JRS4
A0A2W1BNR6
A0A2A4K392
A0A194QXU1
A0A212EUK7
A0A2H1WQC0
+ More
A0A0L7KYQ6 A0A194PMZ2 A0A1B0EU68 D6WBR2 A0A1W4XSU6 N6UMC8 T1HRI1 A0A067QQJ0 A0A2J7RCL4 A0A1B6DR60 A0A336M8R0 A0A1B0D5H1 E0VED2 A0A2P8ZNG4 A0A0M3QWK7 B4J337 B4LFF7 B4L065 A0A0P9C1X1 B3M7T7 B4GR87 M9PCA3 Q9VTW5 Q2M0Q7 A0A0J9RVB3 B4QR90 B4N3J1 A0A3B0JRG4 B4HFL5 A0A0K8W3I9 B4PGC0 A0A0R1DZ32 A0A0L0BYU1 A0A1I8MC98 B3NH92 A0A0Q5UIR0 A0A1J1J714 A0A1A9WD04 A0A1I8QCX9 A0A1W4UUP6 A0A1W4UH56 A0A226EHG3 B0W5E2 A0A2H8TU69 E2BM86 A0A026VZM4 Q17LD3 A0A1B0G3P6 A0A1S4EYJ8 E1ZXG4 A0A1B0BXP5 J9K3G8 A0A1B0A6T7 A0A1A9URU1 A0A195DHV1 F4WKB4 A0A151XAX7 A0A158NQJ4 A0A195D550 A0A0J7KM82 A0A0P6ICS0 A0A0M8ZRP0 A0A1D2NML9 A0A0P6C593 A0A195ERP0 A0A2A3E4R9 A0A195BS25 A0A2S2NHT4 A0A1A9YKR4 A0A182J7Z7 A0A084W9J2 A0A0P5ZX32 K7IQU9 A0A154PN88 A0A182HAX6 U4U621 W5J7V2 A0A1S3H8H1 A0A146LC18 A0A0A9X6B9 A0A1S3JYN4 A0A1S3JXZ8 A0A1B6DS86 A0A182FH98
A0A0L7KYQ6 A0A194PMZ2 A0A1B0EU68 D6WBR2 A0A1W4XSU6 N6UMC8 T1HRI1 A0A067QQJ0 A0A2J7RCL4 A0A1B6DR60 A0A336M8R0 A0A1B0D5H1 E0VED2 A0A2P8ZNG4 A0A0M3QWK7 B4J337 B4LFF7 B4L065 A0A0P9C1X1 B3M7T7 B4GR87 M9PCA3 Q9VTW5 Q2M0Q7 A0A0J9RVB3 B4QR90 B4N3J1 A0A3B0JRG4 B4HFL5 A0A0K8W3I9 B4PGC0 A0A0R1DZ32 A0A0L0BYU1 A0A1I8MC98 B3NH92 A0A0Q5UIR0 A0A1J1J714 A0A1A9WD04 A0A1I8QCX9 A0A1W4UUP6 A0A1W4UH56 A0A226EHG3 B0W5E2 A0A2H8TU69 E2BM86 A0A026VZM4 Q17LD3 A0A1B0G3P6 A0A1S4EYJ8 E1ZXG4 A0A1B0BXP5 J9K3G8 A0A1B0A6T7 A0A1A9URU1 A0A195DHV1 F4WKB4 A0A151XAX7 A0A158NQJ4 A0A195D550 A0A0J7KM82 A0A0P6ICS0 A0A0M8ZRP0 A0A1D2NML9 A0A0P6C593 A0A195ERP0 A0A2A3E4R9 A0A195BS25 A0A2S2NHT4 A0A1A9YKR4 A0A182J7Z7 A0A084W9J2 A0A0P5ZX32 K7IQU9 A0A154PN88 A0A182HAX6 U4U621 W5J7V2 A0A1S3H8H1 A0A146LC18 A0A0A9X6B9 A0A1S3JYN4 A0A1S3JXZ8 A0A1B6DS86 A0A182FH98
Pubmed
19121390
28756777
26354079
22118469
26227816
18362917
+ More
19820115 23537049 24845553 20566863 29403074 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22936249 17550304 26108605 25315136 20798317 24508170 30249741 17510324 21719571 21347285 27289101 24438588 20075255 26483478 20920257 23761445 26823975 25401762
19820115 23537049 24845553 20566863 29403074 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22936249 17550304 26108605 25315136 20798317 24508170 30249741 17510324 21719571 21347285 27289101 24438588 20075255 26483478 20920257 23761445 26823975 25401762
EMBL
BABH01017223
KZ149976
PZC75921.1
NWSH01000237
PCG78122.1
KQ460953
+ More
KPJ10358.1 AGBW02012361 OWR45166.1 ODYU01009905 SOQ54634.1 JTDY01004236 KOB68413.1 KQ459599 KPI94363.1 AJWK01011307 KQ971314 EEZ99306.1 APGK01026589 APGK01026590 KB740635 ENN79872.1 ACPB03000674 KK869568 KDR06680.1 NEVH01005885 PNF38560.1 GEDC01009122 JAS28176.1 UFQT01000253 SSX22428.1 AJVK01025245 AJVK01025246 DS235090 EEB11738.1 PYGN01000010 PSN58019.1 CP012525 ALC44319.1 CH916366 EDV96108.1 CH940647 EDW69255.2 CH933809 EDW18011.2 CH902618 KPU77648.1 EDV38810.1 CH479188 EDW40272.1 AE014296 AGB94447.1 BT003467 AAF49930.2 AAO39470.1 CH379069 EAL30873.2 CM002912 KMY99214.1 CM000363 EDX10220.1 KMY99213.1 CH964095 EDW79196.2 OUUW01000002 SPP76289.1 CH480815 EDW41246.1 GDHF01006660 JAI45654.1 CM000159 EDW94284.1 KRK01742.1 JRES01001238 KNC24404.1 CH954178 EDV51549.1 KQS43846.1 CVRI01000067 CRL06681.1 LNIX01000003 OXA57075.1 DS231842 EDS35262.1 GFXV01005694 MBW17499.1 GL449158 EFN83212.1 KK107648 QOIP01000008 EZA48329.1 RLU19974.1 CH477216 EAT47529.1 CCAG010015899 GL435038 EFN74141.1 JXJN01022295 JXJN01023514 ABLF02035865 ABLF02035867 KQ980824 KYN12478.1 GL888199 EGI65338.1 KQ982335 KYQ57504.1 ADTU01023344 ADTU01023345 ADTU01023346 KQ976885 KYN07554.1 LBMM01005583 KMQ91402.1 GDIQ01006416 JAN88321.1 KQ435922 KOX68586.1 LJIJ01000005 ODN06459.1 GDIP01005670 LRGB01002451 JAM98045.1 KZS07658.1 KQ982021 KYN30544.1 KZ288372 PBC26727.1 KQ976419 KYM89314.1 GGMR01003853 MBY16472.1 ATLV01021825 KE525324 KFB46886.1 GDIP01038505 JAM65210.1 KQ434998 KZC13309.1 JXUM01123612 KQ566736 KXJ69875.1 KB631996 KI210453 ERL87788.1 ERL96272.1 ADMH02002078 ETN59458.1 GDHC01012726 JAQ05903.1 GBHO01029231 JAG14373.1 GEDC01008775 JAS28523.1
KPJ10358.1 AGBW02012361 OWR45166.1 ODYU01009905 SOQ54634.1 JTDY01004236 KOB68413.1 KQ459599 KPI94363.1 AJWK01011307 KQ971314 EEZ99306.1 APGK01026589 APGK01026590 KB740635 ENN79872.1 ACPB03000674 KK869568 KDR06680.1 NEVH01005885 PNF38560.1 GEDC01009122 JAS28176.1 UFQT01000253 SSX22428.1 AJVK01025245 AJVK01025246 DS235090 EEB11738.1 PYGN01000010 PSN58019.1 CP012525 ALC44319.1 CH916366 EDV96108.1 CH940647 EDW69255.2 CH933809 EDW18011.2 CH902618 KPU77648.1 EDV38810.1 CH479188 EDW40272.1 AE014296 AGB94447.1 BT003467 AAF49930.2 AAO39470.1 CH379069 EAL30873.2 CM002912 KMY99214.1 CM000363 EDX10220.1 KMY99213.1 CH964095 EDW79196.2 OUUW01000002 SPP76289.1 CH480815 EDW41246.1 GDHF01006660 JAI45654.1 CM000159 EDW94284.1 KRK01742.1 JRES01001238 KNC24404.1 CH954178 EDV51549.1 KQS43846.1 CVRI01000067 CRL06681.1 LNIX01000003 OXA57075.1 DS231842 EDS35262.1 GFXV01005694 MBW17499.1 GL449158 EFN83212.1 KK107648 QOIP01000008 EZA48329.1 RLU19974.1 CH477216 EAT47529.1 CCAG010015899 GL435038 EFN74141.1 JXJN01022295 JXJN01023514 ABLF02035865 ABLF02035867 KQ980824 KYN12478.1 GL888199 EGI65338.1 KQ982335 KYQ57504.1 ADTU01023344 ADTU01023345 ADTU01023346 KQ976885 KYN07554.1 LBMM01005583 KMQ91402.1 GDIQ01006416 JAN88321.1 KQ435922 KOX68586.1 LJIJ01000005 ODN06459.1 GDIP01005670 LRGB01002451 JAM98045.1 KZS07658.1 KQ982021 KYN30544.1 KZ288372 PBC26727.1 KQ976419 KYM89314.1 GGMR01003853 MBY16472.1 ATLV01021825 KE525324 KFB46886.1 GDIP01038505 JAM65210.1 KQ434998 KZC13309.1 JXUM01123612 KQ566736 KXJ69875.1 KB631996 KI210453 ERL87788.1 ERL96272.1 ADMH02002078 ETN59458.1 GDHC01012726 JAQ05903.1 GBHO01029231 JAG14373.1 GEDC01008775 JAS28523.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000037510
UP000053268
+ More
UP000092461 UP000007266 UP000192223 UP000019118 UP000015103 UP000027135 UP000235965 UP000092462 UP000009046 UP000245037 UP000092553 UP000001070 UP000008792 UP000009192 UP000007801 UP000008744 UP000000803 UP000001819 UP000000304 UP000007798 UP000268350 UP000001292 UP000002282 UP000037069 UP000095301 UP000008711 UP000183832 UP000091820 UP000095300 UP000192221 UP000198287 UP000002320 UP000008237 UP000053097 UP000279307 UP000008820 UP000092444 UP000000311 UP000092460 UP000007819 UP000092445 UP000078200 UP000078492 UP000007755 UP000075809 UP000005205 UP000078542 UP000036403 UP000053105 UP000094527 UP000076858 UP000078541 UP000242457 UP000078540 UP000092443 UP000075880 UP000030765 UP000002358 UP000076502 UP000069940 UP000249989 UP000030742 UP000000673 UP000085678 UP000069272
UP000092461 UP000007266 UP000192223 UP000019118 UP000015103 UP000027135 UP000235965 UP000092462 UP000009046 UP000245037 UP000092553 UP000001070 UP000008792 UP000009192 UP000007801 UP000008744 UP000000803 UP000001819 UP000000304 UP000007798 UP000268350 UP000001292 UP000002282 UP000037069 UP000095301 UP000008711 UP000183832 UP000091820 UP000095300 UP000192221 UP000198287 UP000002320 UP000008237 UP000053097 UP000279307 UP000008820 UP000092444 UP000000311 UP000092460 UP000007819 UP000092445 UP000078200 UP000078492 UP000007755 UP000075809 UP000005205 UP000078542 UP000036403 UP000053105 UP000094527 UP000076858 UP000078541 UP000242457 UP000078540 UP000092443 UP000075880 UP000030765 UP000002358 UP000076502 UP000069940 UP000249989 UP000030742 UP000000673 UP000085678 UP000069272
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
H9JRS4
A0A2W1BNR6
A0A2A4K392
A0A194QXU1
A0A212EUK7
A0A2H1WQC0
+ More
A0A0L7KYQ6 A0A194PMZ2 A0A1B0EU68 D6WBR2 A0A1W4XSU6 N6UMC8 T1HRI1 A0A067QQJ0 A0A2J7RCL4 A0A1B6DR60 A0A336M8R0 A0A1B0D5H1 E0VED2 A0A2P8ZNG4 A0A0M3QWK7 B4J337 B4LFF7 B4L065 A0A0P9C1X1 B3M7T7 B4GR87 M9PCA3 Q9VTW5 Q2M0Q7 A0A0J9RVB3 B4QR90 B4N3J1 A0A3B0JRG4 B4HFL5 A0A0K8W3I9 B4PGC0 A0A0R1DZ32 A0A0L0BYU1 A0A1I8MC98 B3NH92 A0A0Q5UIR0 A0A1J1J714 A0A1A9WD04 A0A1I8QCX9 A0A1W4UUP6 A0A1W4UH56 A0A226EHG3 B0W5E2 A0A2H8TU69 E2BM86 A0A026VZM4 Q17LD3 A0A1B0G3P6 A0A1S4EYJ8 E1ZXG4 A0A1B0BXP5 J9K3G8 A0A1B0A6T7 A0A1A9URU1 A0A195DHV1 F4WKB4 A0A151XAX7 A0A158NQJ4 A0A195D550 A0A0J7KM82 A0A0P6ICS0 A0A0M8ZRP0 A0A1D2NML9 A0A0P6C593 A0A195ERP0 A0A2A3E4R9 A0A195BS25 A0A2S2NHT4 A0A1A9YKR4 A0A182J7Z7 A0A084W9J2 A0A0P5ZX32 K7IQU9 A0A154PN88 A0A182HAX6 U4U621 W5J7V2 A0A1S3H8H1 A0A146LC18 A0A0A9X6B9 A0A1S3JYN4 A0A1S3JXZ8 A0A1B6DS86 A0A182FH98
A0A0L7KYQ6 A0A194PMZ2 A0A1B0EU68 D6WBR2 A0A1W4XSU6 N6UMC8 T1HRI1 A0A067QQJ0 A0A2J7RCL4 A0A1B6DR60 A0A336M8R0 A0A1B0D5H1 E0VED2 A0A2P8ZNG4 A0A0M3QWK7 B4J337 B4LFF7 B4L065 A0A0P9C1X1 B3M7T7 B4GR87 M9PCA3 Q9VTW5 Q2M0Q7 A0A0J9RVB3 B4QR90 B4N3J1 A0A3B0JRG4 B4HFL5 A0A0K8W3I9 B4PGC0 A0A0R1DZ32 A0A0L0BYU1 A0A1I8MC98 B3NH92 A0A0Q5UIR0 A0A1J1J714 A0A1A9WD04 A0A1I8QCX9 A0A1W4UUP6 A0A1W4UH56 A0A226EHG3 B0W5E2 A0A2H8TU69 E2BM86 A0A026VZM4 Q17LD3 A0A1B0G3P6 A0A1S4EYJ8 E1ZXG4 A0A1B0BXP5 J9K3G8 A0A1B0A6T7 A0A1A9URU1 A0A195DHV1 F4WKB4 A0A151XAX7 A0A158NQJ4 A0A195D550 A0A0J7KM82 A0A0P6ICS0 A0A0M8ZRP0 A0A1D2NML9 A0A0P6C593 A0A195ERP0 A0A2A3E4R9 A0A195BS25 A0A2S2NHT4 A0A1A9YKR4 A0A182J7Z7 A0A084W9J2 A0A0P5ZX32 K7IQU9 A0A154PN88 A0A182HAX6 U4U621 W5J7V2 A0A1S3H8H1 A0A146LC18 A0A0A9X6B9 A0A1S3JYN4 A0A1S3JXZ8 A0A1B6DS86 A0A182FH98
PDB
4JCJ
E-value=2.69521e-19,
Score=233
Ontologies
GO
Topology
Subcellular location
Nucleus
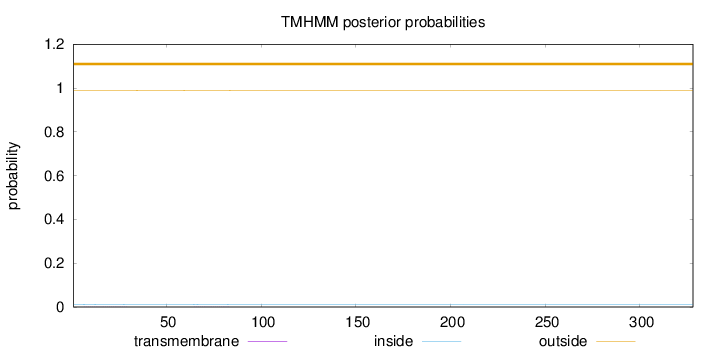
Length:
328
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01542
Exp number, first 60 AAs:
0.00735
Total prob of N-in:
0.01226
outside
1 - 328
Population Genetic Test Statistics
Pi
12.979526
Theta
17.859409
Tajima's D
-0.816499
CLR
0
CSRT
0.175141242937853
Interpretation
Uncertain
