Gene
KWMTBOMO00581
Pre Gene Modal
BGIBMGA012351
Annotation
PREDICTED:_alpha-tocopherol_transfer_protein-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.089 Mitochondrial Reliability : 1.395
Sequence
CDS
ATGTCCAAGAGATTGCAGAAGAGATATCTAACCGAGGCTGACGAGTACAAATGTCCGCTATCCGAGGAGACGCAGACGATAGCTTCAGAAGAGTTGCGGGAGACCGAGAGCGCGAGGACACAGGCTCTGCAGGCCCTGAGGAGTTGGATGGAACAGAACCCTAAGCTGTTGGCTGTTCGACTAGACTCGAATTACTTGCTCCGGTTTCTTCGAGCCAAAAAGTTCAGTGTTCCAATGGCTCAGGAGGCAATCGAACGTTATCTGCTCCTGCGTCAATCCTGGGGCATCGCCTTCAACCAGCTCGACTACAAGCTGCCAATCATGATGGAGCTCTTGGAACTCGGATTCGTTTTCGTCAGCCCCTTCAAGGACAAGGGCGGTCGTCGAGTCGTTATTTATAGACCAGGAGTATTCGACCCTTACAAATACACCAACCAAGACATGTGCCGTGTGATGGGCATCTGCTATGAAACCCTTATGGAAGACGAGGAGACTCAAGTCCGAGGAGTGGTTCACTTCGCTGACGGTGGGGGAGTAGGCTTCCCACATCTCACATTATTCACGCCCAAAGAAGCCGTCCGGATTGTCAAGAATGGAGAGCGCACAGTTCCCGTGAGGCATAAGGAAATTTACGGCATCAATGTCCATCCCACGATCAAGTTCGCTTTAGACTTCGGTATGGCTCTCATATCAGAGAAGATCAGGAAGAGAGTCAGACTTTACACGAGCATAAATGACGTAGAAATAGACCAGAGTTTATTGCCCAAAGAGTACGGCGGAGTTATGCCGATGAAGGAAATGATAGAATTATGGAAAGAGCAGCTCGCGAAAAAACGAGACATGGTACTACTCAATGACAAAATGAGCGTGAGATTAGAGATGTACAGTGAAGCAGCGCGCGAAGGCGCTATCTCCGCTTTAAGATCTGGAGCCAACACGTGCGCTGGGGCAGACGCAGTGGGGGACGCCATGCGAGGTCTCACCGGAAACTTCAGAAAACTTGAAGTCGACTAA
Protein
MSKRLQKRYLTEADEYKCPLSEETQTIASEELRETESARTQALQALRSWMEQNPKLLAVRLDSNYLLRFLRAKKFSVPMAQEAIERYLLLRQSWGIAFNQLDYKLPIMMELLELGFVFVSPFKDKGGRRVVIYRPGVFDPYKYTNQDMCRVMGICYETLMEDEETQVRGVVHFADGGGVGFPHLTLFTPKEAVRIVKNGERTVPVRHKEIYGINVHPTIKFALDFGMALISEKIRKRVRLYTSINDVEIDQSLLPKEYGGVMPMKEMIELWKEQLAKKRDMVLLNDKMSVRLEMYSEAAREGAISALRSGANTCAGADAVGDAMRGLTGNFRKLEVD
Summary
Uniprot
H9JS41
A0A2H1V4I9
A0A2H4RMS6
S4P5B1
A0A2A4JAD1
A0A194QXT4
+ More
A0A212EQF9 I4DLF3 A0A0L7LND3 A0A0N0BJ38 K7IN09 A0A154PAX8 A0A232EUN1 A0A195EGE0 A0A088A3K7 A0A2A3ESN2 A0A195BZV9 A0A151XBM7 F4WG06 A0A195F0M1 A0A195BG52 A0A0J7NHP9 A0A0L7QSH9 A0A158NL16 E2BVA7 A0A3L8D8F9 E2ASK5 D6WH37 A0A1B6FNG7 U4UJG4 A0A026WSH6 A0A1Y1K3A9 A0A1W4X9A7 A0A1Y1K279 A0A182YJN5 N6TQA1 A0A023F3G8 A0A182MHZ6 A0A182W328 A0A182R8F9 A0A224XSK7 A0A182P3G4 A0A182XF13 A0A182VEM5 A0A182TPA3 Q7PQ96 A0A182HVI0 A0A067R5Q1 A0A182SFA0 A0A182GSS2 A0A182Q0E9 A0A336MIP4 A0A182J2L3 A0A2M4BT58 A0A1B6DUL4 A0A2M4BSU0 A0A182FAS6 A0A182NCP0 W5J3E4 A0A1S4FGV5 Q171Y7 A0A2J7QWV1 B0X297 A0A1Q3FQG0 T1I239 A0A2R7WHY3 A0A2H8TNE8 A0A1Q3FQ45 J9JL22 A0A336MXQ0 A0A0T6B116 A0A1B6M111 C4WUM1 A0A1B0DFY3 A0A1B0CH04 A0A3Q0IT56 A0A1J1J4U9 A0A1L8DAJ2 A0A1J1J3N4 A0A1J1J7V7 E0W3B9 A0A1L8DZ54 A0A2J7QIH9 A0A1B0D0M1 A0A182ILW2 A0A1B6IYA5 A0A067RPT1 A0A182PW99 A0A069DTA5 A0A2P8XVF4 A0A182NM49 A0A1B6HID8 A0A182FCH8
A0A212EQF9 I4DLF3 A0A0L7LND3 A0A0N0BJ38 K7IN09 A0A154PAX8 A0A232EUN1 A0A195EGE0 A0A088A3K7 A0A2A3ESN2 A0A195BZV9 A0A151XBM7 F4WG06 A0A195F0M1 A0A195BG52 A0A0J7NHP9 A0A0L7QSH9 A0A158NL16 E2BVA7 A0A3L8D8F9 E2ASK5 D6WH37 A0A1B6FNG7 U4UJG4 A0A026WSH6 A0A1Y1K3A9 A0A1W4X9A7 A0A1Y1K279 A0A182YJN5 N6TQA1 A0A023F3G8 A0A182MHZ6 A0A182W328 A0A182R8F9 A0A224XSK7 A0A182P3G4 A0A182XF13 A0A182VEM5 A0A182TPA3 Q7PQ96 A0A182HVI0 A0A067R5Q1 A0A182SFA0 A0A182GSS2 A0A182Q0E9 A0A336MIP4 A0A182J2L3 A0A2M4BT58 A0A1B6DUL4 A0A2M4BSU0 A0A182FAS6 A0A182NCP0 W5J3E4 A0A1S4FGV5 Q171Y7 A0A2J7QWV1 B0X297 A0A1Q3FQG0 T1I239 A0A2R7WHY3 A0A2H8TNE8 A0A1Q3FQ45 J9JL22 A0A336MXQ0 A0A0T6B116 A0A1B6M111 C4WUM1 A0A1B0DFY3 A0A1B0CH04 A0A3Q0IT56 A0A1J1J4U9 A0A1L8DAJ2 A0A1J1J3N4 A0A1J1J7V7 E0W3B9 A0A1L8DZ54 A0A2J7QIH9 A0A1B0D0M1 A0A182ILW2 A0A1B6IYA5 A0A067RPT1 A0A182PW99 A0A069DTA5 A0A2P8XVF4 A0A182NM49 A0A1B6HID8 A0A182FCH8
Pubmed
EMBL
BABH01017216
ODYU01000641
SOQ35758.1
MG434637
ATY51943.1
GAIX01005649
+ More
JAA86911.1 NWSH01002345 PCG68484.1 KQ460953 KPJ10287.1 AGBW02013270 OWR43701.1 AK402121 KQ459605 BAM18743.1 KPI92024.1 JTDY01000466 KOB77053.1 KQ435724 KOX78448.1 KQ434846 KZC08378.1 NNAY01002105 OXU22069.1 KQ978957 KYN26959.1 KZ288187 PBC34710.1 KQ978501 KYM93461.1 KQ982320 KYQ57781.1 GL888128 EGI66773.1 KQ981866 KYN34130.1 KQ976488 KYM83572.1 LBMM01004861 KMQ92030.1 KQ414758 KOC61504.1 ADTU01019141 GL450824 EFN80399.1 QOIP01000011 RLU16373.1 GL442330 EFN63565.1 KQ971321 EFA00115.1 GECZ01018014 JAS51755.1 KB632330 ERL92608.1 KK107111 EZA58933.1 GEZM01096816 JAV54560.1 GEZM01096815 JAV54561.1 APGK01055647 APGK01055648 KB741269 ENN71420.1 GBBI01002886 JAC15826.1 AXCM01000316 GFTR01004914 JAW11512.1 AAAB01008898 EAA09146.6 APCN01005778 KK852680 KDR18600.1 JXUM01017356 KQ560481 KXJ82198.1 AXCN02001182 UFQT01001063 SSX28703.1 GGFJ01007109 MBW56250.1 GEDC01007937 JAS29361.1 GGFJ01007015 MBW56156.1 ADMH02002202 ETN57843.1 CH477443 EAT40799.1 NEVH01009418 PNF33062.1 DS232283 EDS39120.1 GFDL01005317 JAV29728.1 ACPB03000289 KK854855 PTY19253.1 GFXV01003754 MBW15559.1 GFDL01005358 JAV29687.1 ABLF02028481 UFQT01003484 SSX35046.1 LJIG01016378 KRT80782.1 GEBQ01010364 JAT29613.1 AK341126 BAH71591.1 AJVK01059119 AJWK01011791 AJWK01011792 AJWK01011793 AJWK01011794 CVRI01000069 CRL06986.1 GFDF01010724 JAV03360.1 CRL06984.1 CRL06985.1 DS235882 EEB20125.1 GFDF01002346 JAV11738.1 NEVH01013592 PNF28391.1 AJVK01009964 AJVK01009965 GECU01015789 JAS91917.1 KK852542 KDR21724.1 GBGD01002017 JAC86872.1 PYGN01001288 PSN35983.1 GECU01033278 JAS74428.1
JAA86911.1 NWSH01002345 PCG68484.1 KQ460953 KPJ10287.1 AGBW02013270 OWR43701.1 AK402121 KQ459605 BAM18743.1 KPI92024.1 JTDY01000466 KOB77053.1 KQ435724 KOX78448.1 KQ434846 KZC08378.1 NNAY01002105 OXU22069.1 KQ978957 KYN26959.1 KZ288187 PBC34710.1 KQ978501 KYM93461.1 KQ982320 KYQ57781.1 GL888128 EGI66773.1 KQ981866 KYN34130.1 KQ976488 KYM83572.1 LBMM01004861 KMQ92030.1 KQ414758 KOC61504.1 ADTU01019141 GL450824 EFN80399.1 QOIP01000011 RLU16373.1 GL442330 EFN63565.1 KQ971321 EFA00115.1 GECZ01018014 JAS51755.1 KB632330 ERL92608.1 KK107111 EZA58933.1 GEZM01096816 JAV54560.1 GEZM01096815 JAV54561.1 APGK01055647 APGK01055648 KB741269 ENN71420.1 GBBI01002886 JAC15826.1 AXCM01000316 GFTR01004914 JAW11512.1 AAAB01008898 EAA09146.6 APCN01005778 KK852680 KDR18600.1 JXUM01017356 KQ560481 KXJ82198.1 AXCN02001182 UFQT01001063 SSX28703.1 GGFJ01007109 MBW56250.1 GEDC01007937 JAS29361.1 GGFJ01007015 MBW56156.1 ADMH02002202 ETN57843.1 CH477443 EAT40799.1 NEVH01009418 PNF33062.1 DS232283 EDS39120.1 GFDL01005317 JAV29728.1 ACPB03000289 KK854855 PTY19253.1 GFXV01003754 MBW15559.1 GFDL01005358 JAV29687.1 ABLF02028481 UFQT01003484 SSX35046.1 LJIG01016378 KRT80782.1 GEBQ01010364 JAT29613.1 AK341126 BAH71591.1 AJVK01059119 AJWK01011791 AJWK01011792 AJWK01011793 AJWK01011794 CVRI01000069 CRL06986.1 GFDF01010724 JAV03360.1 CRL06984.1 CRL06985.1 DS235882 EEB20125.1 GFDF01002346 JAV11738.1 NEVH01013592 PNF28391.1 AJVK01009964 AJVK01009965 GECU01015789 JAS91917.1 KK852542 KDR21724.1 GBGD01002017 JAC86872.1 PYGN01001288 PSN35983.1 GECU01033278 JAS74428.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000053105 UP000002358 UP000076502 UP000215335 UP000078492 UP000005203 UP000242457 UP000078542 UP000075809 UP000007755 UP000078541 UP000078540 UP000036403 UP000053825 UP000005205 UP000008237 UP000279307 UP000000311 UP000007266 UP000030742 UP000053097 UP000192223 UP000076408 UP000019118 UP000075883 UP000075920 UP000075900 UP000075885 UP000076407 UP000075903 UP000075902 UP000007062 UP000075840 UP000027135 UP000075901 UP000069940 UP000249989 UP000075886 UP000075880 UP000069272 UP000075884 UP000000673 UP000008820 UP000235965 UP000002320 UP000015103 UP000007819 UP000092462 UP000092461 UP000079169 UP000183832 UP000009046 UP000245037
UP000053105 UP000002358 UP000076502 UP000215335 UP000078492 UP000005203 UP000242457 UP000078542 UP000075809 UP000007755 UP000078541 UP000078540 UP000036403 UP000053825 UP000005205 UP000008237 UP000279307 UP000000311 UP000007266 UP000030742 UP000053097 UP000192223 UP000076408 UP000019118 UP000075883 UP000075920 UP000075900 UP000075885 UP000076407 UP000075903 UP000075902 UP000007062 UP000075840 UP000027135 UP000075901 UP000069940 UP000249989 UP000075886 UP000075880 UP000069272 UP000075884 UP000000673 UP000008820 UP000235965 UP000002320 UP000015103 UP000007819 UP000092462 UP000092461 UP000079169 UP000183832 UP000009046 UP000245037
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JS41
A0A2H1V4I9
A0A2H4RMS6
S4P5B1
A0A2A4JAD1
A0A194QXT4
+ More
A0A212EQF9 I4DLF3 A0A0L7LND3 A0A0N0BJ38 K7IN09 A0A154PAX8 A0A232EUN1 A0A195EGE0 A0A088A3K7 A0A2A3ESN2 A0A195BZV9 A0A151XBM7 F4WG06 A0A195F0M1 A0A195BG52 A0A0J7NHP9 A0A0L7QSH9 A0A158NL16 E2BVA7 A0A3L8D8F9 E2ASK5 D6WH37 A0A1B6FNG7 U4UJG4 A0A026WSH6 A0A1Y1K3A9 A0A1W4X9A7 A0A1Y1K279 A0A182YJN5 N6TQA1 A0A023F3G8 A0A182MHZ6 A0A182W328 A0A182R8F9 A0A224XSK7 A0A182P3G4 A0A182XF13 A0A182VEM5 A0A182TPA3 Q7PQ96 A0A182HVI0 A0A067R5Q1 A0A182SFA0 A0A182GSS2 A0A182Q0E9 A0A336MIP4 A0A182J2L3 A0A2M4BT58 A0A1B6DUL4 A0A2M4BSU0 A0A182FAS6 A0A182NCP0 W5J3E4 A0A1S4FGV5 Q171Y7 A0A2J7QWV1 B0X297 A0A1Q3FQG0 T1I239 A0A2R7WHY3 A0A2H8TNE8 A0A1Q3FQ45 J9JL22 A0A336MXQ0 A0A0T6B116 A0A1B6M111 C4WUM1 A0A1B0DFY3 A0A1B0CH04 A0A3Q0IT56 A0A1J1J4U9 A0A1L8DAJ2 A0A1J1J3N4 A0A1J1J7V7 E0W3B9 A0A1L8DZ54 A0A2J7QIH9 A0A1B0D0M1 A0A182ILW2 A0A1B6IYA5 A0A067RPT1 A0A182PW99 A0A069DTA5 A0A2P8XVF4 A0A182NM49 A0A1B6HID8 A0A182FCH8
A0A212EQF9 I4DLF3 A0A0L7LND3 A0A0N0BJ38 K7IN09 A0A154PAX8 A0A232EUN1 A0A195EGE0 A0A088A3K7 A0A2A3ESN2 A0A195BZV9 A0A151XBM7 F4WG06 A0A195F0M1 A0A195BG52 A0A0J7NHP9 A0A0L7QSH9 A0A158NL16 E2BVA7 A0A3L8D8F9 E2ASK5 D6WH37 A0A1B6FNG7 U4UJG4 A0A026WSH6 A0A1Y1K3A9 A0A1W4X9A7 A0A1Y1K279 A0A182YJN5 N6TQA1 A0A023F3G8 A0A182MHZ6 A0A182W328 A0A182R8F9 A0A224XSK7 A0A182P3G4 A0A182XF13 A0A182VEM5 A0A182TPA3 Q7PQ96 A0A182HVI0 A0A067R5Q1 A0A182SFA0 A0A182GSS2 A0A182Q0E9 A0A336MIP4 A0A182J2L3 A0A2M4BT58 A0A1B6DUL4 A0A2M4BSU0 A0A182FAS6 A0A182NCP0 W5J3E4 A0A1S4FGV5 Q171Y7 A0A2J7QWV1 B0X297 A0A1Q3FQG0 T1I239 A0A2R7WHY3 A0A2H8TNE8 A0A1Q3FQ45 J9JL22 A0A336MXQ0 A0A0T6B116 A0A1B6M111 C4WUM1 A0A1B0DFY3 A0A1B0CH04 A0A3Q0IT56 A0A1J1J4U9 A0A1L8DAJ2 A0A1J1J3N4 A0A1J1J7V7 E0W3B9 A0A1L8DZ54 A0A2J7QIH9 A0A1B0D0M1 A0A182ILW2 A0A1B6IYA5 A0A067RPT1 A0A182PW99 A0A069DTA5 A0A2P8XVF4 A0A182NM49 A0A1B6HID8 A0A182FCH8
PDB
3W68
E-value=6.8579e-16,
Score=204
Ontologies
GO
Topology
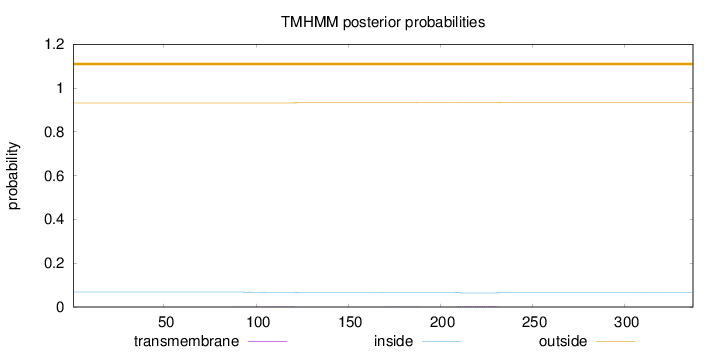
Length:
337
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11342
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.06810
outside
1 - 337
Population Genetic Test Statistics
Pi
23.682357
Theta
24.683765
Tajima's D
-1.119004
CLR
0.022542
CSRT
0.114344282785861
Interpretation
Uncertain
