Gene
KWMTBOMO00578 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012239
Annotation
PREDICTED:_uncharacterized_protein_LOC101745012_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.819
Sequence
CDS
ATGGAGGCAGAGGACGTGGCAGACAATGAATTCTTGGAAGTTAAGCCCAAAGATCTCGGTCTTGATGGACATCTACGACGACTGAAACGTGGTCTATTTGACAGTTGGTTTTCGCCTTTTGACACGTCTTCGACTACTGAGGCGCCCCCTGAAAACATCGAGTCGATCACTGAAGACGATGCGAGCTTCGACAGGAAAGACGACGATTTGGACACCGAAGGCTCCGGCTACCCGGAGAACAGAGTCGAGCCAGAAATTAAAGAAAAGACCCTTCGTGTTACCTTCGTAGTCATGGAACCATATCGAGAAGAGTACGTCAACAGAGATTCGATACAGTTCCAGAACTTATCCAAATCTCTTGCTGATGAAGTCAATAAGCTCTTCGAATCTTTGCCTGGCACACAAAAGGCTAGCCTAGTGAGAATCCAGTCTTGCGTCTCCGACGACTTCTCCTGCAAAGTTACTCTGGACATTGTAACAACTGGATACGACAACACGGACGTGATCGCTCAAACCCTGAGAGACCACATCAGGAACCGAAGCATGCTAGGGAAGGTCACAGTAAACGATAACGATTTCAGCGCTTTTGTTATTGATTCAGCAAACTTCACTTGTCGCCCTAATGAGATCCAGTGCGGCGACGGTGCTTGCGTCCCAGAATCGGCAAGATGTAATAATGAAAGGGACTGTAGCGATTGGTCCGACGAAAAGAACTGCCCTATTGAAAACATTCCCGATGAGGAACAAGAACAGACCTTAGAGGCTGAAACAGAAAGTTATCGTCCTATCGAACGTGGCGATCAGCCCTTCGCATCTGAAACGACCACGCCTTATTATGACGAAGACGAAGTTACAGTTCCCTATCCTGAACAATTTACATGTTCATTGGACGAGTTCAAATGCGATGAGACCCGTTGCATCAGACGGGACGAGGTGTGCAACAACATCAACGACTGTCACGATGGAACTGACGAAGTGGACTGCGCAAACCTTGACCACAAGATGGCATTTGAGTTGTACATGTGGCCTCCTCGAGTCCCTTAA
Protein
MEAEDVADNEFLEVKPKDLGLDGHLRRLKRGLFDSWFSPFDTSSTTEAPPENIESITEDDASFDRKDDDLDTEGSGYPENRVEPEIKEKTLRVTFVVMEPYREEYVNRDSIQFQNLSKSLADEVNKLFESLPGTQKASLVRIQSCVSDDFSCKVTLDIVTTGYDNTDVIAQTLRDHIRNRSMLGKVTVNDNDFSAFVIDSANFTCRPNEIQCGDGACVPESARCNNERDCSDWSDEKNCPIENIPDEEQEQTLEAETESYRPIERGDQPFASETTTPYYDEDEVTVPYPEQFTCSLDEFKCDETRCIRRDEVCNNINDCHDGTDEVDCANLDHKMAFELYMWPPRVP
Summary
Uniprot
H9JRS9
A0A1E1W5F9
A0A1E1VXS1
A0A194PG74
A0A194QZ87
A0A139WI18
+ More
A0A139WI17 A0A139WIN8 A0A139WIA3 A0A1Y1MVG2 A0A1Y1N267 A0A1V1FQN6 A0A087ZY95 A0A1B6H0Y4 A0A1B6EZM0 A0A1B6FYY0 A0A195AUX2 A0A224XSJ6 A0A0L7QT60 A0A151WH30 A0A195FWA5 E9IR36 A0A158NND7 A0A195BZ34 A0A310SDI3 A0A1B6KPM4 A0A026X1H8 F7CCA0 A0A3L8DHC7 A0A1B6HNG0 A0A2A3EF83 A0A1B6KBT2 A0A1L8FP59 A0A1B6IQI8 A0A1B6IHQ9 A0A1B6J9M1 A0A0M9A216 I3KII9 A0A1W5AS91 A0A087T6N4 A0A3Q0IR92 I3KIJ0 A0A2I4CIQ1 A0A3L8RXT3 A0A3B4C5S2 A0A2U9C270
A0A139WI17 A0A139WIN8 A0A139WIA3 A0A1Y1MVG2 A0A1Y1N267 A0A1V1FQN6 A0A087ZY95 A0A1B6H0Y4 A0A1B6EZM0 A0A1B6FYY0 A0A195AUX2 A0A224XSJ6 A0A0L7QT60 A0A151WH30 A0A195FWA5 E9IR36 A0A158NND7 A0A195BZ34 A0A310SDI3 A0A1B6KPM4 A0A026X1H8 F7CCA0 A0A3L8DHC7 A0A1B6HNG0 A0A2A3EF83 A0A1B6KBT2 A0A1L8FP59 A0A1B6IQI8 A0A1B6IHQ9 A0A1B6J9M1 A0A0M9A216 I3KII9 A0A1W5AS91 A0A087T6N4 A0A3Q0IR92 I3KIJ0 A0A2I4CIQ1 A0A3L8RXT3 A0A3B4C5S2 A0A2U9C270
Pubmed
EMBL
BABH01017209
GDQN01008875
JAT82179.1
GDQN01011583
JAT79471.1
KQ459605
+ More
KPI92028.1 KQ460953 KPJ10290.1 KQ971342 KYB27640.1 KYB27638.1 KYB27637.1 KYB27639.1 GEZM01019399 JAV89661.1 GEZM01019402 JAV89657.1 FX985450 BAX07463.1 GECZ01001452 JAS68317.1 GECZ01026263 JAS43506.1 GECZ01014383 JAS55386.1 KQ976738 KYM75825.1 GFTR01004924 JAW11502.1 KQ414756 KOC61661.1 KQ983136 KYQ47141.1 KQ981208 KYN44706.1 GL765043 EFZ16969.1 ADTU01021233 ADTU01021234 ADTU01021235 ADTU01021236 ADTU01021237 ADTU01021238 ADTU01021239 ADTU01021240 ADTU01021241 ADTU01021242 KQ978457 KYM93872.1 KQ773953 OAD52335.1 GEBQ01026592 JAT13385.1 KK107031 EZA62102.1 AAMC01127504 AAMC01127505 AAMC01127506 AAMC01127507 AAMC01127508 AAMC01127509 AAMC01127510 AAMC01127511 AAMC01127512 QOIP01000008 RLU19726.1 GECU01031496 JAS76210.1 KZ288266 PBC30144.1 GEBQ01031102 JAT08875.1 CM004478 OCT73376.1 GECU01018549 JAS89157.1 GECU01021259 JAS86447.1 GECU01011877 JAS95829.1 KQ435760 KOX75587.1 AERX01002528 AERX01002529 KK113670 KFM60773.1 QUSF01000139 RLV89744.1 CP026253 AWP10153.1
KPI92028.1 KQ460953 KPJ10290.1 KQ971342 KYB27640.1 KYB27638.1 KYB27637.1 KYB27639.1 GEZM01019399 JAV89661.1 GEZM01019402 JAV89657.1 FX985450 BAX07463.1 GECZ01001452 JAS68317.1 GECZ01026263 JAS43506.1 GECZ01014383 JAS55386.1 KQ976738 KYM75825.1 GFTR01004924 JAW11502.1 KQ414756 KOC61661.1 KQ983136 KYQ47141.1 KQ981208 KYN44706.1 GL765043 EFZ16969.1 ADTU01021233 ADTU01021234 ADTU01021235 ADTU01021236 ADTU01021237 ADTU01021238 ADTU01021239 ADTU01021240 ADTU01021241 ADTU01021242 KQ978457 KYM93872.1 KQ773953 OAD52335.1 GEBQ01026592 JAT13385.1 KK107031 EZA62102.1 AAMC01127504 AAMC01127505 AAMC01127506 AAMC01127507 AAMC01127508 AAMC01127509 AAMC01127510 AAMC01127511 AAMC01127512 QOIP01000008 RLU19726.1 GECU01031496 JAS76210.1 KZ288266 PBC30144.1 GEBQ01031102 JAT08875.1 CM004478 OCT73376.1 GECU01018549 JAS89157.1 GECU01021259 JAS86447.1 GECU01011877 JAS95829.1 KQ435760 KOX75587.1 AERX01002528 AERX01002529 KK113670 KFM60773.1 QUSF01000139 RLV89744.1 CP026253 AWP10153.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007266
UP000005203
UP000078540
+ More
UP000053825 UP000075809 UP000078541 UP000005205 UP000078542 UP000053097 UP000008143 UP000279307 UP000242457 UP000186698 UP000053105 UP000005207 UP000192224 UP000054359 UP000079169 UP000192220 UP000276834 UP000261440 UP000246464
UP000053825 UP000075809 UP000078541 UP000005205 UP000078542 UP000053097 UP000008143 UP000279307 UP000242457 UP000186698 UP000053105 UP000005207 UP000192224 UP000054359 UP000079169 UP000192220 UP000276834 UP000261440 UP000246464
PRIDE
Pfam
Interpro
IPR023415
LDLR_class-A_CS
+ More
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR000082 SEA_dom
IPR036364 SEA_dom_sf
IPR003598 Ig_sub2
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR036179 Ig-like_dom_sf
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR000034 Laminin_IV
IPR002049 Laminin_EGF
IPR013151 Immunoglobulin
IPR013098 Ig_I-set
IPR009030 Growth_fac_rcpt_cys_sf
IPR001881 EGF-like_Ca-bd_dom
IPR013320 ConA-like_dom_sf
IPR001791 Laminin_G
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR013106 Ig_V-set
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR000082 SEA_dom
IPR036364 SEA_dom_sf
IPR003598 Ig_sub2
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR036179 Ig-like_dom_sf
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR000034 Laminin_IV
IPR002049 Laminin_EGF
IPR013151 Immunoglobulin
IPR013098 Ig_I-set
IPR009030 Growth_fac_rcpt_cys_sf
IPR001881 EGF-like_Ca-bd_dom
IPR013320 ConA-like_dom_sf
IPR001791 Laminin_G
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR013106 Ig_V-set
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9JRS9
A0A1E1W5F9
A0A1E1VXS1
A0A194PG74
A0A194QZ87
A0A139WI18
+ More
A0A139WI17 A0A139WIN8 A0A139WIA3 A0A1Y1MVG2 A0A1Y1N267 A0A1V1FQN6 A0A087ZY95 A0A1B6H0Y4 A0A1B6EZM0 A0A1B6FYY0 A0A195AUX2 A0A224XSJ6 A0A0L7QT60 A0A151WH30 A0A195FWA5 E9IR36 A0A158NND7 A0A195BZ34 A0A310SDI3 A0A1B6KPM4 A0A026X1H8 F7CCA0 A0A3L8DHC7 A0A1B6HNG0 A0A2A3EF83 A0A1B6KBT2 A0A1L8FP59 A0A1B6IQI8 A0A1B6IHQ9 A0A1B6J9M1 A0A0M9A216 I3KII9 A0A1W5AS91 A0A087T6N4 A0A3Q0IR92 I3KIJ0 A0A2I4CIQ1 A0A3L8RXT3 A0A3B4C5S2 A0A2U9C270
A0A139WI17 A0A139WIN8 A0A139WIA3 A0A1Y1MVG2 A0A1Y1N267 A0A1V1FQN6 A0A087ZY95 A0A1B6H0Y4 A0A1B6EZM0 A0A1B6FYY0 A0A195AUX2 A0A224XSJ6 A0A0L7QT60 A0A151WH30 A0A195FWA5 E9IR36 A0A158NND7 A0A195BZ34 A0A310SDI3 A0A1B6KPM4 A0A026X1H8 F7CCA0 A0A3L8DHC7 A0A1B6HNG0 A0A2A3EF83 A0A1B6KBT2 A0A1L8FP59 A0A1B6IQI8 A0A1B6IHQ9 A0A1B6J9M1 A0A0M9A216 I3KII9 A0A1W5AS91 A0A087T6N4 A0A3Q0IR92 I3KIJ0 A0A2I4CIQ1 A0A3L8RXT3 A0A3B4C5S2 A0A2U9C270
PDB
6BYV
E-value=6.82493e-08,
Score=135
Ontologies
GO
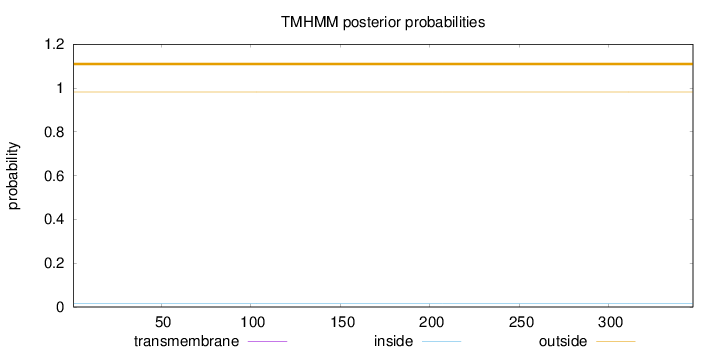
Topology
Length:
347
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01769
outside
1 - 347
Population Genetic Test Statistics
Pi
25.864924
Theta
26.50685
Tajima's D
0.077424
CLR
0.050093
CSRT
0.395530223488826
Interpretation
Uncertain
