Gene
KWMTBOMO00576 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012240
Annotation
PREDICTED:_uncharacterized_protein_LOC101745012_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.999
Sequence
CDS
ATGATATCTATTACAGCAAAATCATGTGGTATCGACGACTTTACTTGCAACAACGGAGCCTGTATCGAGGGTTTCAGACGTTGTGATGGCGCTGTGGACTGTTCTGAAGGCGAAGACGAAGTTAATTGTGAATGCAGAGAGGACCAATTCAGGTGTAGATCTGGATCATGTATTGAATTGAGAAAACGCTGTGATGGATACGAACACTGCAGCGACCGCTCAGACGAGGAGAACTGCAACGGTACGTTGAGTTGTCCACCAGAGAAGTTCGCTTGTAGTCCAACATCATCGGTGCCGTGTGCTATAAGATGCAATGGCTTCCCCGAATGCGATGGGAACGAAGACGAAGACAACTGTCCAGACGAAGAATGCAGCCACCAGTGCGACGGGAAATGTCTTCCGGATGCGAATATCTGTGACTCTATCACGGACTGTTCAGACGGTTCGGACGAGTTGCATTGTGCACAGTGCGATGGTCCAGACGATTACAGGTGTGATACTGACGAGTGTATAAGCAGAAATTGGGTTTGTAACGGCCACTTCGAATGTCCTAACGGCGAAGACGAGGCAGACTGCAACAGTACTATAGTTTCGACTCCAACATGTGACGAAAACAGCTGGCAGTGCCGAAGCGGTCAATGCATACCGTCCAGCAGCTATTGCGACAACGTCTACAACTGTTACGATAGGTCGGATGAAGCCGATTGCCTTCCATCAACTCCTGCATACGAAAACGAATACCCACCAATAGTTTCTACAACTTTGAGTCCTTACTACACAACGACTCGACGAGATGTTGGATCCAGCAATTATGACAGAAGACCTGAAAACCCATACGGATCGGAAAACCAAGGGACATATTCGCCTTACGACGACGACGCAAGACGTCCTACAACGTATTCACCATACGGACCCAGTTCAGAGAGACCTGATCCGCGTCGACCGTATCCACCATATGGCTCTGTAACTACCAACCGACCCGATGACAGGCAGGATCCCTATAGCAGCCCACCTGGTGGGGAATATGATCCTTATGGACAAGGTGCTGGTAGCCCAGATGATAGACGTCCTCCATACAGTCAACCTGGAAATCAAGACATCAGTCCTGGAGGTGGAAGCCCATACAATCCTGAACCAGAAAACCAACCGGATAATAGAGAAGATCCCTCTGGCGGCCAACCTGGCGGTGAGTATAATCCTTATGGCCAGGGTGCTGGTGGACCAGACGATAGACGTCCACCGTATACTCAACCTGGAAGTCAAGATGTAAGTCCAGGAGGTGGAAGCCCATACAATCCCGAATCAGGAAGTCAACCAAATAACAGAGAAGATCCTTATGGTGGCCAGTCTGGTGGTGAATATGATCCTTATGGCCGGGGTGCTGGTAGTCCAGATGATAGACGTCCTCCATACGGTCAATCTGGAAGCCAAGACATCAGTCTTGGAGGTGGAATACCATACAACCCCGAATCAGTAAGGCAACCAGACAACAGAGATCCCTATGGCGGCCAATCCGGTGGTGAATATGATCCCTATGGGCGGGGCGTAAGTGGAACAGACGACAGACGTCAACCTGGAAGCCAAGATGTTGGTCCAGGAGCTTCGGGTCAAGGCAATTCAAATCAACCAGAAAACCGGCCCTATCGTCCTGAAAGTGGCAATAGCGTTTATGAGACGGGTGAGGGGCAATATGGATCTGGTAGTCAAAGAGGAGACAGAGGTCCCATTCCGTTAAATATCAAAACATATCCGCAAGACCAGGAATCTGTCAGATACGGTGGTGATGTAGTGCTTCAGTGCCGTGATGAAGGCCCTCGTCGTGCACCCGTGAGATGGATCCGAGAAGGTGGTAGGCCGATGAGGCCAAACTACCAGGAAAGAAAAGGACGACTGGAACTGTTTGGTGTTACCCCATCCGATAGTGGTGTATACATATGTCAGGCACCATCTTATCTGAGACAGCCTGGTGCGGAAGTTCGAGTTACTTTGGAAGTGGAGACGAGCCCTGTGACCGTGCGGCCAGTGTTCGGTGTTTGTAAGCCACATGAAGCCACTTGCGGTAACGGGCAGTGCATACCAAAATCGGCTATTTGCAACTCTGTTTTCGACTGCGCTGATCATTCAGACGAAGATGGCTGCGGAGCCAGTGGCCAGTGCGAACCGAACGAGTTTGTTTGCGCGAACCACAAGTGCATTCTTAAGACCTGGCGTTGTGACTCCGAGGACGACTGCGGCGATGGTTCTGATGAACTTAACTGTGGAACCCCAACTCCTGGAAGCCCTTGCTTGGCGGTAGAATTCTCATGTGCTTCTAACAACCAATGTATTCCGAAGAGCTTCCACTGTGACGGACAATCTGACTGCGTTGATGGCTCTGATGAAATCGGATGCGCTCAAGTATATATCACGAAACCCCCGCAACCTTCATACGTGAGATTGAATCCGGGCGACTCGTTGACTCTAAGATGTGAGGCCGTTGGTGTACCTGTTCCACTAGTCTCGTGGCGTCTCAACTGGGGACATGTTCCACCTCAATGCACCTTTACCAGTGACAACGGTATCGGAGTTTTGACATGTCCAAACATGCAGCCCCAACATAGCGGTGCGTATTCGTGCGAGGCCATCAATAATAAGGCTACGACATTCGCGACACCGGACGCAATCGTACACGTGAATAGAACCGATCCTTGCCCCGTTGGATATTTCAATAGCGATGCGCGTTCACAAAGCGAATGCATCCAGTGCTTCTGCTTCGGCGAGTCCACGCAGTGTCGCAGCGCGGATCTGTTTACGTACAACATGCCGACTCCTCTTGGAGAAGGCGGCACCAGATTCGTCACTGTCACTCATCAGGCCAGAGGCGACATCAGTATCGGCGCCCCGATGAACAACGAATACTTCTATCAACCTTTGAGGAACGGAGCTACTGTGACCAGAATAGTCCCCGGAGACATGGGCTGGTTCAAAGATGCTCATCCATACGTGACCTTACCAGAAACCTACAACGGTAACCAGTTAACTTCATACGGTGGTCACATCAAATACACCATCTCGCCTCACGGATCCGGATATGGCTCAGATGACACTGCACCAACCATAATCATCAAGGGGAAATACTTCACTCTGTTCCATTACTACCGCGGCGAGACGACTAAAACTATCAACATCGAAGCTCGTTTGACTCCCGAGAACTGGCTGATTCTGGACGAAAACGGCGTTCAAGCGCCAGCGCAGAGATCGACCATCATGATGACGTAG
Protein
MISITAKSCGIDDFTCNNGACIEGFRRCDGAVDCSEGEDEVNCECREDQFRCRSGSCIELRKRCDGYEHCSDRSDEENCNGTLSCPPEKFACSPTSSVPCAIRCNGFPECDGNEDEDNCPDEECSHQCDGKCLPDANICDSITDCSDGSDELHCAQCDGPDDYRCDTDECISRNWVCNGHFECPNGEDEADCNSTIVSTPTCDENSWQCRSGQCIPSSSYCDNVYNCYDRSDEADCLPSTPAYENEYPPIVSTTLSPYYTTTRRDVGSSNYDRRPENPYGSENQGTYSPYDDDARRPTTYSPYGPSSERPDPRRPYPPYGSVTTNRPDDRQDPYSSPPGGEYDPYGQGAGSPDDRRPPYSQPGNQDISPGGGSPYNPEPENQPDNREDPSGGQPGGEYNPYGQGAGGPDDRRPPYTQPGSQDVSPGGGSPYNPESGSQPNNREDPYGGQSGGEYDPYGRGAGSPDDRRPPYGQSGSQDISLGGGIPYNPESVRQPDNRDPYGGQSGGEYDPYGRGVSGTDDRRQPGSQDVGPGASGQGNSNQPENRPYRPESGNSVYETGEGQYGSGSQRGDRGPIPLNIKTYPQDQESVRYGGDVVLQCRDEGPRRAPVRWIREGGRPMRPNYQERKGRLELFGVTPSDSGVYICQAPSYLRQPGAEVRVTLEVETSPVTVRPVFGVCKPHEATCGNGQCIPKSAICNSVFDCADHSDEDGCGASGQCEPNEFVCANHKCILKTWRCDSEDDCGDGSDELNCGTPTPGSPCLAVEFSCASNNQCIPKSFHCDGQSDCVDGSDEIGCAQVYITKPPQPSYVRLNPGDSLTLRCEAVGVPVPLVSWRLNWGHVPPQCTFTSDNGIGVLTCPNMQPQHSGAYSCEAINNKATTFATPDAIVHVNRTDPCPVGYFNSDARSQSECIQCFCFGESTQCRSADLFTYNMPTPLGEGGTRFVTVTHQARGDISIGAPMNNEYFYQPLRNGATVTRIVPGDMGWFKDAHPYVTLPETYNGNQLTSYGGHIKYTISPHGSGYGSDDTAPTIIIKGKYFTLFHYYRGETTKTINIEARLTPENWLILDENGVQAPAQRSTIMMT
Summary
Uniprot
Pubmed
Proteomes
PRIDE
Pfam
Interpro
IPR001791
Laminin_G
+ More
IPR000742 EGF-like_dom
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013032 EGF-like_CS
IPR023415 LDLR_class-A_CS
IPR013320 ConA-like_dom_sf
IPR013151 Immunoglobulin
IPR007110 Ig-like_dom
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR001881 EGF-like_Ca-bd_dom
IPR000034 Laminin_IV
IPR013783 Ig-like_fold
IPR002049 Laminin_EGF
IPR003599 Ig_sub
IPR009030 Growth_fac_rcpt_cys_sf
IPR000742 EGF-like_dom
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013032 EGF-like_CS
IPR023415 LDLR_class-A_CS
IPR013320 ConA-like_dom_sf
IPR013151 Immunoglobulin
IPR007110 Ig-like_dom
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR001881 EGF-like_Ca-bd_dom
IPR000034 Laminin_IV
IPR013783 Ig-like_fold
IPR002049 Laminin_EGF
IPR003599 Ig_sub
IPR009030 Growth_fac_rcpt_cys_sf
Gene 3D
CDD
ProteinModelPortal
PDB
3M0C
E-value=2.56043e-27,
Score=308
Ontologies
GO
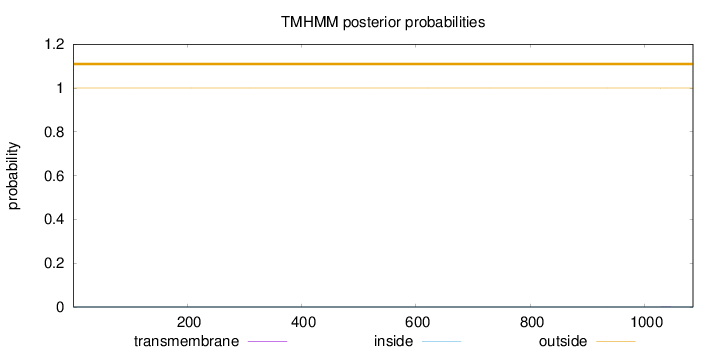
Topology
Length:
1085
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00361
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00000
outside
1 - 1085
Population Genetic Test Statistics
Pi
20.518479
Theta
21.771417
Tajima's D
0.006942
CLR
0.117785
CSRT
0.380580970951452
Interpretation
Uncertain
