Gene
KWMTBOMO00556
Pre Gene Modal
BGIBMGA012343
Annotation
PREDICTED:_tyrosine-protein_phosphatase_corkscrew-like_[Amyelois_transitella]
Full name
Tyrosine-protein phosphatase non-receptor type
Location in the cell
Nuclear Reliability : 3.021
Sequence
CDS
ATGTCGAAAGATAATCCAAGGTTGAGCCGGCTCGGAGTGTTGGCGCCGGCTGCCCTGTCGGCCTGCATGGATGCCTCCGTGCTCGCAGTCCCGGCTGCAGGTGCAATCGTCTCCACGGTTCTCAAGGAGTTGAGCAAGGCCTTCATGCTCAAAAAATGGTTCCACGGTGTTATGTCAGCCAAAGAAGCCGAGAGCCTCATCATGGAGAAAGGGAAGAACGGTTCGTTCTTGGTCCGAGAATCTCAGGCGCATCCCGGTGAATACGTGCTGTCTGTCAGAGTCAGAGGTCGAGTTAGCCACGTCCTTATCAGACGACAGCACAACAAATACGACGTAGGTAGCGGTGAGCAGTTCGATGACCTGGTGGGTCTTATTGAACACTTTCGTTCGTATCCGATGATCGAGACATCAGGAGACGTACTGCGTCTCTTGCAGCCTGTTAGTGGCACCAGACTAAGAGCTCACGACATCGACCAGAAAGTACAGGAGATGCAGGGAATAGAAGATCGACAAAAAATGGAATGTAATACCGGTTTCGATGGTGAGTTTCAGTCATTGAAACTGCTTGAAGACAGACACGTGTTCACAACTATCGAGGGAATGAAGCCTGAGAACGTTTCGAAGAACAGATATAGAAATGTATTGCCTTATGACCAAACGCGGGTAGTTCTTCGTCGTCGAGATAACAAACTGGAATCGGACTACATCAACGCATGCTTCGTACGATCGTCGCGTCTCAGTGATTCTACAAGCTCTGTCCAGTCATCGAACGAAAGTCTTGACAGTGTTCAATCGTTGATCCTGAATACTGAAAAGAAGAAGGCAGTACCACTCGTCACGAAATCGCTTTCAGACGACGCTCTGAGAGAAATGAAGAAAAGTCTTAAATTAGATAAAATTAATGGGAATCTAAATAAAGATGCAGTAAAAGACAAAATATATATCGCCGCTCAGGGATGTTTGCCGAGCACTAAGGATGACTTCTGGCGAATGATCTGGCAGGAGGACGTCAGAGTTATAGCGATGATCACGAACGAACATGAAAAGGGGAAGAGAAAATGTGAACGTTATTGGCCATTATCGGGACAGGAAGAGCGATATGACAACCTCCTTGTGAGATCCATTTCTGAGTCGCACTACGAAGATTACCTAATTAGGGAGCTTGACGTATGCGACGAAAATAAATGTTGCAGAGCAGTTTATCAATATCAGTTTACGGCCTGGCCGGACCACAGTACTCCACCGGAGCCGGACAGGATAATCTCATTTATGGAGGATATAAACAGGAGAATGTTTCTAATCACACAAGAGAAAAATGCACCAGATCAGAATGTGATTTGCGTGCATTGTTCCGCGGGAGTCGGGAGAACCGGCACGTTCATAGTCTTGGACATCCTTCTTGACAAAATCAAATCATCTGGTTTTAACTGTGACATCGACGTACACAATACAGTGAAGCTAGTGAGGGCTCAGAGAAGCGGCATGGTACAGAATAAAACCCAATATAGATTCATATATCTCGCCCTTCAGGATTACATTGAAAACAAAAATATTAAATTGAGACGGAAGGGCTACCGGCCGGACATCTGTCGCGACTTCCATCACTAA
Protein
MSKDNPRLSRLGVLAPAALSACMDASVLAVPAAGAIVSTVLKELSKAFMLKKWFHGVMSAKEAESLIMEKGKNGSFLVRESQAHPGEYVLSVRVRGRVSHVLIRRQHNKYDVGSGEQFDDLVGLIEHFRSYPMIETSGDVLRLLQPVSGTRLRAHDIDQKVQEMQGIEDRQKMECNTGFDGEFQSLKLLEDRHVFTTIEGMKPENVSKNRYRNVLPYDQTRVVLRRRDNKLESDYINACFVRSSRLSDSTSSVQSSNESLDSVQSLILNTEKKKAVPLVTKSLSDDALREMKKSLKLDKINGNLNKDAVKDKIYIAAQGCLPSTKDDFWRMIWQEDVRVIAMITNEHEKGKRKCERYWPLSGQEERYDNLLVRSISESHYEDYLIRELDVCDENKCCRAVYQYQFTAWPDHSTPPEPDRIISFMEDINRRMFLITQEKNAPDQNVICVHCSAGVGRTGTFIVLDILLDKIKSSGFNCDIDVHNTVKLVRAQRSGMVQNKTQYRFIYLALQDYIENKNIKLRRKGYRPDICRDFHH
Summary
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Similarity
Belongs to the protein-tyrosine phosphatase family. Non-receptor class 2 subfamily.
Uniprot
A0A212EQI6
A0A2A4JVZ2
A0A2W1BB59
H9JS33
A0A2A4JVP3
A0A2H1WQZ9
+ More
A0A2A4JVA9 A0A2A4JU47 A0A194QXU9 A0A194RJU9 A0A194Q681 H9IV20 A0A2H1WY52 A0A2A4K380 A0A0L7LLZ9 V5G2H4 A0A195DM14 A0A154PEJ5 D7ELF2 A0A182JH09 V5GJE6 A0A195F3Y8 A0A1J1HPS2 A0A1J1HV72 A0A1Q3FY24 A0A1J1HR93 A0A182ULX7 A0A182LF66 A0A067QLL8 U4U270 A0A182NFH2 Q5TTR3 J3JX41 A0A2R5L5Q5 U4V0R5 A0A084VU59 N6TKK0 A0A1S3HFG4 A0A293LFB0 A0A0A1XPL0 A0A0A1WVB4 A0A1Q3FXY5 A0A1Q3FV42 A0A1Q3FV66 A0A3B5AD19 A0A3B4F223 I3KDR2 A0A3P8R4H9 A0A3P9BLI1 A0A3P8SUJ1 E0VXC8 A0A3B4AX11 T1JTX1 F6WVD2 A0A3B4VQC1 G9KJA0 A0A151NSB1 A0A218V6D8 H0Z8W9 U3K2T1 A0A3Q0G8L8 A0A0T6B2D1 U5ER21 M7B1N1 G3PSG4 F5HK48 A0A182HT30 G3PSG6 A0A182XI48 A0A3B4XDF9 A0A182TBD5 A0A182MX54 A0A094NHH7 A0A0R3T0Z4 A0A151N4C4 A0A3Q0G3B0 A0A0A0ASV2 A0A091LSF2 A0A3B1J9Z1 A0A093L6F5 A0A1L1RU24 A0A093P6M0 A0A226PF42 A0A2I0M3D4 U3J954 A0A0Q3LZE7 A0A226N0A8 R0KFJ5 A0A0R3X2L0 A0A2P4TGI2 A0A1V4KNV7 U6JGQ4 G1MWD5 A0A0R3VSG3 A0A1A8CU23 G1P7X2
A0A2A4JVA9 A0A2A4JU47 A0A194QXU9 A0A194RJU9 A0A194Q681 H9IV20 A0A2H1WY52 A0A2A4K380 A0A0L7LLZ9 V5G2H4 A0A195DM14 A0A154PEJ5 D7ELF2 A0A182JH09 V5GJE6 A0A195F3Y8 A0A1J1HPS2 A0A1J1HV72 A0A1Q3FY24 A0A1J1HR93 A0A182ULX7 A0A182LF66 A0A067QLL8 U4U270 A0A182NFH2 Q5TTR3 J3JX41 A0A2R5L5Q5 U4V0R5 A0A084VU59 N6TKK0 A0A1S3HFG4 A0A293LFB0 A0A0A1XPL0 A0A0A1WVB4 A0A1Q3FXY5 A0A1Q3FV42 A0A1Q3FV66 A0A3B5AD19 A0A3B4F223 I3KDR2 A0A3P8R4H9 A0A3P9BLI1 A0A3P8SUJ1 E0VXC8 A0A3B4AX11 T1JTX1 F6WVD2 A0A3B4VQC1 G9KJA0 A0A151NSB1 A0A218V6D8 H0Z8W9 U3K2T1 A0A3Q0G8L8 A0A0T6B2D1 U5ER21 M7B1N1 G3PSG4 F5HK48 A0A182HT30 G3PSG6 A0A182XI48 A0A3B4XDF9 A0A182TBD5 A0A182MX54 A0A094NHH7 A0A0R3T0Z4 A0A151N4C4 A0A3Q0G3B0 A0A0A0ASV2 A0A091LSF2 A0A3B1J9Z1 A0A093L6F5 A0A1L1RU24 A0A093P6M0 A0A226PF42 A0A2I0M3D4 U3J954 A0A0Q3LZE7 A0A226N0A8 R0KFJ5 A0A0R3X2L0 A0A2P4TGI2 A0A1V4KNV7 U6JGQ4 G1MWD5 A0A0R3VSG3 A0A1A8CU23 G1P7X2
EC Number
3.1.3.48
Pubmed
22118469
28756777
19121390
26354079
26227816
18362917
+ More
19820115 20966253 24845553 23537049 12364791 22516182 24438588 25830018 25186727 20566863 25463417 17495919 23236062 22293439 20360741 23624526 14747013 17210077 25329095 15592404 24621616 23371554 23749191 24013640 23485966 20838655 21993624
19820115 20966253 24845553 23537049 12364791 22516182 24438588 25830018 25186727 20566863 25463417 17495919 23236062 22293439 20360741 23624526 14747013 17210077 25329095 15592404 24621616 23371554 23749191 24013640 23485966 20838655 21993624
EMBL
AGBW02013269
OWR43760.1
NWSH01000575
PCG75552.1
KZ150203
PZC72268.1
+ More
BABH01017146 PCG75553.1 ODYU01010354 SOQ55406.1 PCG75554.1 PCG75555.1 KQ460953 KPJ10302.1 KQ460297 KPJ16226.1 KQ459439 KPJ01043.1 BABH01000164 ODYU01011528 SOQ57314.1 NWSH01000210 PCG78378.1 JTDY01000610 KOB76472.1 GALX01004251 JAB64215.1 KQ980762 KYN13534.1 KQ434889 KZC10247.1 DS497846 EFA12064.1 GALX01004252 JAB64214.1 KQ981826 KYN35106.1 CVRI01000015 CRK90056.1 CRK90057.1 GFDL01002632 JAV32413.1 CRK90058.1 KK853191 KDR10028.1 KB632006 ERL87959.1 AAAB01008859 EAL40926.4 BT127809 AEE62771.1 GGLE01000677 MBY04803.1 KI210464 ERL96276.1 ATLV01016641 ATLV01016642 ATLV01016643 ATLV01016644 KE525099 KFB41503.1 APGK01034121 APGK01034122 KB740904 ENN78448.1 GFWV01001828 MAA26558.1 GBXI01001512 JAD12780.1 GBXI01011711 JAD02581.1 GFDL01002679 JAV32366.1 GFDL01003723 JAV31322.1 GFDL01003722 JAV31323.1 AERX01036520 AERX01036521 DS235830 EEB18034.1 CAEY01000486 JP016381 AES04979.1 AKHW03002185 KYO39678.1 MUZQ01000045 OWK61142.1 ABQF01028897 ABQF01028898 ABQF01028899 ABQF01028900 AGTO01020434 LJIG01016140 KRT81496.1 GANO01003972 JAB55899.1 KB585326 EMP25998.1 EGK96889.1 APCN01000403 AXCM01003184 KL361808 KFZ65951.1 UZAE01000147 VDN96378.1 AKHW03004073 KYO31537.1 KL872854 KGL96947.1 KK509203 KFP62453.1 KK598797 KFW04891.1 AADN05000197 KL225141 KFW67902.1 AWGT02000089 OXB78182.1 AKCR02000044 PKK24176.1 ADON01000006 ADON01000007 LMAW01002900 KQK75864.1 MCFN01000301 OXB61007.1 KB742382 EOB09216.1 UYWX01020379 VDM31911.1 PPHD01000442 POI35466.1 LSYS01002427 OPJ86103.1 LK028589 APAU02000020 CDS23237.1 EUB61468.1 UYRS01000008 VDK20229.1 HADZ01018260 SBP82201.1 AAPE02037827 AAPE02037828
BABH01017146 PCG75553.1 ODYU01010354 SOQ55406.1 PCG75554.1 PCG75555.1 KQ460953 KPJ10302.1 KQ460297 KPJ16226.1 KQ459439 KPJ01043.1 BABH01000164 ODYU01011528 SOQ57314.1 NWSH01000210 PCG78378.1 JTDY01000610 KOB76472.1 GALX01004251 JAB64215.1 KQ980762 KYN13534.1 KQ434889 KZC10247.1 DS497846 EFA12064.1 GALX01004252 JAB64214.1 KQ981826 KYN35106.1 CVRI01000015 CRK90056.1 CRK90057.1 GFDL01002632 JAV32413.1 CRK90058.1 KK853191 KDR10028.1 KB632006 ERL87959.1 AAAB01008859 EAL40926.4 BT127809 AEE62771.1 GGLE01000677 MBY04803.1 KI210464 ERL96276.1 ATLV01016641 ATLV01016642 ATLV01016643 ATLV01016644 KE525099 KFB41503.1 APGK01034121 APGK01034122 KB740904 ENN78448.1 GFWV01001828 MAA26558.1 GBXI01001512 JAD12780.1 GBXI01011711 JAD02581.1 GFDL01002679 JAV32366.1 GFDL01003723 JAV31322.1 GFDL01003722 JAV31323.1 AERX01036520 AERX01036521 DS235830 EEB18034.1 CAEY01000486 JP016381 AES04979.1 AKHW03002185 KYO39678.1 MUZQ01000045 OWK61142.1 ABQF01028897 ABQF01028898 ABQF01028899 ABQF01028900 AGTO01020434 LJIG01016140 KRT81496.1 GANO01003972 JAB55899.1 KB585326 EMP25998.1 EGK96889.1 APCN01000403 AXCM01003184 KL361808 KFZ65951.1 UZAE01000147 VDN96378.1 AKHW03004073 KYO31537.1 KL872854 KGL96947.1 KK509203 KFP62453.1 KK598797 KFW04891.1 AADN05000197 KL225141 KFW67902.1 AWGT02000089 OXB78182.1 AKCR02000044 PKK24176.1 ADON01000006 ADON01000007 LMAW01002900 KQK75864.1 MCFN01000301 OXB61007.1 KB742382 EOB09216.1 UYWX01020379 VDM31911.1 PPHD01000442 POI35466.1 LSYS01002427 OPJ86103.1 LK028589 APAU02000020 CDS23237.1 EUB61468.1 UYRS01000008 VDK20229.1 HADZ01018260 SBP82201.1 AAPE02037827 AAPE02037828
Proteomes
UP000007151
UP000218220
UP000005204
UP000053240
UP000053268
UP000037510
+ More
UP000078492 UP000076502 UP000007266 UP000075880 UP000078541 UP000183832 UP000075903 UP000075882 UP000027135 UP000030742 UP000075884 UP000007062 UP000030765 UP000019118 UP000085678 UP000261400 UP000261460 UP000005207 UP000265100 UP000265160 UP000265080 UP000009046 UP000261520 UP000015104 UP000002280 UP000261420 UP000050525 UP000197619 UP000007754 UP000016665 UP000189705 UP000031443 UP000007635 UP000075840 UP000076407 UP000261360 UP000075901 UP000075883 UP000046398 UP000278807 UP000053858 UP000018467 UP000000539 UP000054081 UP000198419 UP000053872 UP000016666 UP000051836 UP000198323 UP000046396 UP000274429 UP000190648 UP000019149 UP000001645 UP000046400 UP000282613 UP000001074
UP000078492 UP000076502 UP000007266 UP000075880 UP000078541 UP000183832 UP000075903 UP000075882 UP000027135 UP000030742 UP000075884 UP000007062 UP000030765 UP000019118 UP000085678 UP000261400 UP000261460 UP000005207 UP000265100 UP000265160 UP000265080 UP000009046 UP000261520 UP000015104 UP000002280 UP000261420 UP000050525 UP000197619 UP000007754 UP000016665 UP000189705 UP000031443 UP000007635 UP000075840 UP000076407 UP000261360 UP000075901 UP000075883 UP000046398 UP000278807 UP000053858 UP000018467 UP000000539 UP000054081 UP000198419 UP000053872 UP000016666 UP000051836 UP000198323 UP000046396 UP000274429 UP000190648 UP000019149 UP000001645 UP000046400 UP000282613 UP000001074
Interpro
Gene 3D
ProteinModelPortal
A0A212EQI6
A0A2A4JVZ2
A0A2W1BB59
H9JS33
A0A2A4JVP3
A0A2H1WQZ9
+ More
A0A2A4JVA9 A0A2A4JU47 A0A194QXU9 A0A194RJU9 A0A194Q681 H9IV20 A0A2H1WY52 A0A2A4K380 A0A0L7LLZ9 V5G2H4 A0A195DM14 A0A154PEJ5 D7ELF2 A0A182JH09 V5GJE6 A0A195F3Y8 A0A1J1HPS2 A0A1J1HV72 A0A1Q3FY24 A0A1J1HR93 A0A182ULX7 A0A182LF66 A0A067QLL8 U4U270 A0A182NFH2 Q5TTR3 J3JX41 A0A2R5L5Q5 U4V0R5 A0A084VU59 N6TKK0 A0A1S3HFG4 A0A293LFB0 A0A0A1XPL0 A0A0A1WVB4 A0A1Q3FXY5 A0A1Q3FV42 A0A1Q3FV66 A0A3B5AD19 A0A3B4F223 I3KDR2 A0A3P8R4H9 A0A3P9BLI1 A0A3P8SUJ1 E0VXC8 A0A3B4AX11 T1JTX1 F6WVD2 A0A3B4VQC1 G9KJA0 A0A151NSB1 A0A218V6D8 H0Z8W9 U3K2T1 A0A3Q0G8L8 A0A0T6B2D1 U5ER21 M7B1N1 G3PSG4 F5HK48 A0A182HT30 G3PSG6 A0A182XI48 A0A3B4XDF9 A0A182TBD5 A0A182MX54 A0A094NHH7 A0A0R3T0Z4 A0A151N4C4 A0A3Q0G3B0 A0A0A0ASV2 A0A091LSF2 A0A3B1J9Z1 A0A093L6F5 A0A1L1RU24 A0A093P6M0 A0A226PF42 A0A2I0M3D4 U3J954 A0A0Q3LZE7 A0A226N0A8 R0KFJ5 A0A0R3X2L0 A0A2P4TGI2 A0A1V4KNV7 U6JGQ4 G1MWD5 A0A0R3VSG3 A0A1A8CU23 G1P7X2
A0A2A4JVA9 A0A2A4JU47 A0A194QXU9 A0A194RJU9 A0A194Q681 H9IV20 A0A2H1WY52 A0A2A4K380 A0A0L7LLZ9 V5G2H4 A0A195DM14 A0A154PEJ5 D7ELF2 A0A182JH09 V5GJE6 A0A195F3Y8 A0A1J1HPS2 A0A1J1HV72 A0A1Q3FY24 A0A1J1HR93 A0A182ULX7 A0A182LF66 A0A067QLL8 U4U270 A0A182NFH2 Q5TTR3 J3JX41 A0A2R5L5Q5 U4V0R5 A0A084VU59 N6TKK0 A0A1S3HFG4 A0A293LFB0 A0A0A1XPL0 A0A0A1WVB4 A0A1Q3FXY5 A0A1Q3FV42 A0A1Q3FV66 A0A3B5AD19 A0A3B4F223 I3KDR2 A0A3P8R4H9 A0A3P9BLI1 A0A3P8SUJ1 E0VXC8 A0A3B4AX11 T1JTX1 F6WVD2 A0A3B4VQC1 G9KJA0 A0A151NSB1 A0A218V6D8 H0Z8W9 U3K2T1 A0A3Q0G8L8 A0A0T6B2D1 U5ER21 M7B1N1 G3PSG4 F5HK48 A0A182HT30 G3PSG6 A0A182XI48 A0A3B4XDF9 A0A182TBD5 A0A182MX54 A0A094NHH7 A0A0R3T0Z4 A0A151N4C4 A0A3Q0G3B0 A0A0A0ASV2 A0A091LSF2 A0A3B1J9Z1 A0A093L6F5 A0A1L1RU24 A0A093P6M0 A0A226PF42 A0A2I0M3D4 U3J954 A0A0Q3LZE7 A0A226N0A8 R0KFJ5 A0A0R3X2L0 A0A2P4TGI2 A0A1V4KNV7 U6JGQ4 G1MWD5 A0A0R3VSG3 A0A1A8CU23 G1P7X2
PDB
2B3O
E-value=1.31783e-75,
Score=721
Ontologies
GO
GO:0004725
GO:0005737
GO:0060027
GO:0003146
GO:0060548
GO:0060028
GO:0048703
GO:0016740
GO:0036302
GO:0032991
GO:0046326
GO:0001784
GO:0046676
GO:0042445
GO:0071364
GO:0060125
GO:0070374
GO:0048806
GO:0032755
GO:0004726
GO:0060020
GO:0007229
GO:0021697
GO:0048873
GO:0005634
GO:0007173
GO:0005158
GO:0061582
GO:0048013
GO:0035855
GO:0000187
GO:0051463
GO:0060325
GO:0043254
GO:0046887
GO:0033277
GO:0051428
GO:0035264
GO:0032728
GO:0009755
GO:0048609
GO:0045931
GO:0030971
GO:0050839
GO:0005070
GO:0046628
GO:0048008
GO:0033629
GO:0032760
GO:0035265
GO:0000077
GO:0032528
GO:0048011
GO:0030220
GO:0042593
GO:0006641
GO:0048839
GO:0007409
GO:0046825
GO:0006470
GO:0016311
GO:0016791
GO:0035023
GO:0005515
GO:0019752
GO:0030246
GO:0007169
GO:0016021
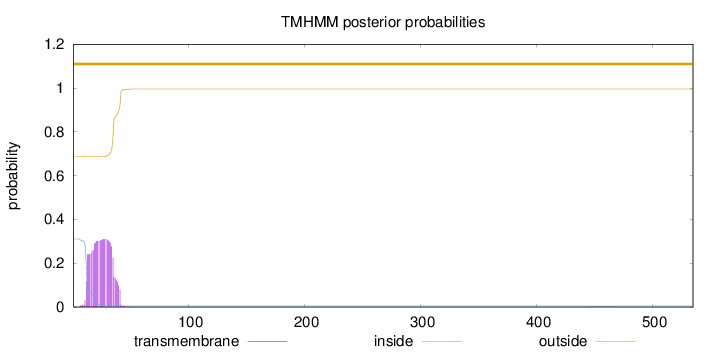
Topology
Subcellular location
Cytoplasm
Length:
535
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.41588999999999
Exp number, first 60 AAs:
7.40737
Total prob of N-in:
0.31107
outside
1 - 535
Population Genetic Test Statistics
Pi
27.349102
Theta
26.88763
Tajima's D
0.178211
CLR
0.036805
CSRT
0.425778711064447
Interpretation
Uncertain
