Gene
KWMTBOMO00555
Pre Gene Modal
BGIBMGA012340
Annotation
PREDICTED:_sodium-dependent_nutrient_amino_acid_transporter_1-like_[Bombyx_mori]
Full name
Transporter
Location in the cell
PlasmaMembrane Reliability : 4.867
Sequence
CDS
ATGTTTGATACCTATAAATTAAAGAAATGGATCAGTTGGCCGTTGATTCGTGCGCACTTATGCACCGTAGCAGTTGCAGTGGGTTTTACGAGCACATGGCGTGCGCCTCGAGAAGCATTCCGTTACGGCGGTCTAACATTCGCTCTGGTATTGACACTGGCATTCGTGGTGGTAGCTTTGCCGGTTGTTTTGCTACAGTTGGCTATTGGACAGCTCAGCCAGCAAGATGCCGTTGGAGTCTGGAGAGCAGTGCCGTTCTTTAAGGGTGTTGGTTATCTCCGGTTGTTGGTTACGACTTTGAGCGCAATATACACGAGTGTTTATTTATCACTAGTCATAACATATTCTTTGTACACCCTAAGTAATAATATTCCCTTCCCGGAGTGTATGGGATTCTATATTACACCGGAAATATATGTGAATGCCATGAACTCTTCAGAATGCTTAGATGAAACATTTCTAGCACCTGCAAGCGAAAAACCAGAGTATTTTTTGGCAGTGGCGCTGATTCTTATTGTACTATGGATACTGTTTCCGTTTATATTTTACAACCCGGTAAAATTTATGAAACGCATATTCTACGCACTCGGGCCCCTGATAGTATTGGTGACGTTCATAATTTTGATCGCGCACGGGGATAAAGGCAATTTGTCATCGTTCGCTCGGTCCAGTGACTGGATGAACTTTCTGCGTCCTGATATTTGGCATGGGGCTATCGTTCAAGCGTTGATATCAAGTCAGGTCGCGGGAGGGTTCCTCATATCTGCTGGAGATTCCGTTTACTCCAGCACGAACGTTCAGTGGACGGCTGTAGCTATCGTTGGCGCCAACCTATTAGCATGTTGGATAGGCATGATATTCTGGTACAGTGTTACTGGTCCTGTCAAACACACAGGAGTGCTCGCAGTTCTCGAGCAAGTATATCAAACAGCAGAAGATGACCAACTGAATAAAACGTGGACGCTACTCGTATTTGCATTGTTGTTTTTATCAGGAGTTGTTTCTACGCTAGCATTTCTTTATCCGGTTTACGATAGACTACGGCGGCTGGGAGGCCTAAAGTGGAGAGTCACTTCGATAGCGTGTTCATTGCTCGGGGCGTGTCTGTCTGTGGCGACATTGGCGGGCCGGCTACCGGCGTTGACTCTTATTGAAGACACGGCTACACCCATGTTGATAAGCTTGACTACTATTCTAGAGATACTACCTTTTATTTTTATTTACGGATGGAAACTTTTAGTGGAAGATGTAGAATTCCTCATCGGTGCCGAGCTGGTGAGATACTGGGTTTGGGGCTGGTGTGCGGCGCCGGGAGTCATTGCTCCGATGTTCGTGTGGTGGCTTACTGTCAATTCTATGGACGTTTGGGAATGGAACCAGCCACCTTGGTCAGCCAGTACTGCTTCAGTAATCGCGGTTCTAGCATTACTCATCTTCGCTATAGCCGCCGCAGTATCTGTGAAAAAACAAGTGCAGTTTGACTTTTTCGGGAAACTGCATTCCTCGTTCGTGCCATCCAGACAGTGGGGTCCCAGAGATCCGATCACTCACTACTACTGGTTGGCGTGCCGCGAGGAGACAGACCGTGGCAATGTTCCTCGATGCCGATACCAAAGACGACCACTAGGTCAACTCTCAACCAGACCCAGCTTCTTAGTTGTGCCCAATACACCACTGAAGATGGAAACAAAGCAAAGATCGAATTCTGATGATTGGTTGTACACCACTAACAGGCGAAAATATTTATCTCGCATTGTCGAACGTTACGTGGGCACCCGGAAACGGTCCAAGTCCTTGGATTGGGATGTTGCTGGAGCAGCAAAAAAATATTTAAACGTTAGTTTAGATTCCATTCTTAGTGTCAACAATAATGATGTAGTATTTAAGGGCGCTTCGGAAAAAGTACTCAAATAA
Protein
MFDTYKLKKWISWPLIRAHLCTVAVAVGFTSTWRAPREAFRYGGLTFALVLTLAFVVVALPVVLLQLAIGQLSQQDAVGVWRAVPFFKGVGYLRLLVTTLSAIYTSVYLSLVITYSLYTLSNNIPFPECMGFYITPEIYVNAMNSSECLDETFLAPASEKPEYFLAVALILIVLWILFPFIFYNPVKFMKRIFYALGPLIVLVTFIILIAHGDKGNLSSFARSSDWMNFLRPDIWHGAIVQALISSQVAGGFLISAGDSVYSSTNVQWTAVAIVGANLLACWIGMIFWYSVTGPVKHTGVLAVLEQVYQTAEDDQLNKTWTLLVFALLFLSGVVSTLAFLYPVYDRLRRLGGLKWRVTSIACSLLGACLSVATLAGRLPALTLIEDTATPMLISLTTILEILPFIFIYGWKLLVEDVEFLIGAELVRYWVWGWCAAPGVIAPMFVWWLTVNSMDVWEWNQPPWSASTASVIAVLALLIFAIAAAVSVKKQVQFDFFGKLHSSFVPSRQWGPRDPITHYYWLACREETDRGNVPRCRYQRRPLGQLSTRPSFLVVPNTPLKMETKQRSNSDDWLYTTNRRKYLSRIVERYVGTRKRSKSLDWDVAGAAKKYLNVSLDSILSVNNNDVVFKGASEKVLK
Summary
Similarity
Belongs to the sodium:neurotransmitter symporter (SNF) (TC 2.A.22) family.
Uniprot
A0A2H1WUH2
A0A2A4JU52
A0A2A4JUW4
A0A2W1BHG8
A0A194PFI0
A0A212EQK9
+ More
A0A194R384 A0A194PIN1 A0A194QNJ1 A0A2H1VKR8 A0A2A4JQ39 H9JLX6 A0A2H8TMM4 X2D4J3 A0A146MEM0 A0A146LQK9 A0A2S2PQU7 A0A1Y1MI14 A0A1Y1ML21 J9KES3 A0A1Y1MIE3 A0A2S2PMX6 A0A2S2QZ54 A0A069DWH7 T1HU77 D6WED3 B6CY69 N6U4N6 A0A1B6DBA5 A0A1B6CRS2 A0A2J7Q3G4 A0A3Q0IT57 A0A1B6LCC7 W8AYE8 B4KZ22 A0A1B6F5R4 A0A0Q9XPD1 A0A1B6GDB9 A0A0M3QWZ4 A0A0A1WZX2 A0A0K8UXH4 A0A0R1E314 A0A1W4W4W9 A0A0Q5UJS4 A0A0K8UVD8 Q9VVG3 Q8IQQ6 A0A0P8YGX5 B4J247 C7LAC7 A0A1W4W4B2 B4PJW1 A0A0J9UN26 B3NHW0 B3M6C3 B4QNU2 B4HKR7 Q8IQQ7 C7LA83 A0A0J9RYC3 Q9U975 A0A3B0KG05 A0A1A9VYS9 B5DQ44 B4MMQ9 A0A034VAZ8 A0A1L8DJV5 A0A1L8DJV8 A0A1A9Y1H6 A0A1B0C1B4 A0A1B0FAG9 A0A1I8MQX4 B4H6L6 A0A1B0C4T3 A0A1A9WEB0 A0A1I8PP83 A0A1Q3EXX7 A0A310SK53 B0WT87 A0A336MYS6 A0A1S4FRX2 A0A1I8PP26 A0A182G8F8 W5JQB4 A0A1Q3EXK4 A0A182FEN9 Q16RP7 A0A1Q3EXK9 A0A1Q3EXU0 A0A182T2I6 A0A2M4BEJ5 A0A182THX6 A0A2M4BDW0 A0A1B0EY28 A0A2M3YYZ3
A0A194R384 A0A194PIN1 A0A194QNJ1 A0A2H1VKR8 A0A2A4JQ39 H9JLX6 A0A2H8TMM4 X2D4J3 A0A146MEM0 A0A146LQK9 A0A2S2PQU7 A0A1Y1MI14 A0A1Y1ML21 J9KES3 A0A1Y1MIE3 A0A2S2PMX6 A0A2S2QZ54 A0A069DWH7 T1HU77 D6WED3 B6CY69 N6U4N6 A0A1B6DBA5 A0A1B6CRS2 A0A2J7Q3G4 A0A3Q0IT57 A0A1B6LCC7 W8AYE8 B4KZ22 A0A1B6F5R4 A0A0Q9XPD1 A0A1B6GDB9 A0A0M3QWZ4 A0A0A1WZX2 A0A0K8UXH4 A0A0R1E314 A0A1W4W4W9 A0A0Q5UJS4 A0A0K8UVD8 Q9VVG3 Q8IQQ6 A0A0P8YGX5 B4J247 C7LAC7 A0A1W4W4B2 B4PJW1 A0A0J9UN26 B3NHW0 B3M6C3 B4QNU2 B4HKR7 Q8IQQ7 C7LA83 A0A0J9RYC3 Q9U975 A0A3B0KG05 A0A1A9VYS9 B5DQ44 B4MMQ9 A0A034VAZ8 A0A1L8DJV5 A0A1L8DJV8 A0A1A9Y1H6 A0A1B0C1B4 A0A1B0FAG9 A0A1I8MQX4 B4H6L6 A0A1B0C4T3 A0A1A9WEB0 A0A1I8PP83 A0A1Q3EXX7 A0A310SK53 B0WT87 A0A336MYS6 A0A1S4FRX2 A0A1I8PP26 A0A182G8F8 W5JQB4 A0A1Q3EXK4 A0A182FEN9 Q16RP7 A0A1Q3EXK9 A0A1Q3EXU0 A0A182T2I6 A0A2M4BEJ5 A0A182THX6 A0A2M4BDW0 A0A1B0EY28 A0A2M3YYZ3
Pubmed
28756777
26354079
22118469
19121390
26823975
28004739
+ More
26334808 18362917 19820115 18621706 23537049 24495485 17994087 18057021 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 10433833 15632085 25348373 25315136 26483478 20920257 23761445 17510324
26334808 18362917 19820115 18621706 23537049 24495485 17994087 18057021 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 10433833 15632085 25348373 25315136 26483478 20920257 23761445 17510324
EMBL
ODYU01011147
SOQ56711.1
NWSH01000575
PCG75557.1
PCG75556.1
KZ150203
+ More
PZC72266.1 KQ459605 KPI92042.1 AGBW02013269 OWR43759.1 KQ460953 KPJ10306.1 KQ459603 KPI92908.1 KQ461194 KPJ06924.1 ODYU01003081 SOQ41391.1 NWSH01000927 PCG73520.1 BABH01005368 BABH01005369 BABH01005370 GFXV01003570 MBW15375.1 KC713794 AHH29252.1 GDHC01001187 JAQ17442.1 GDHC01009060 JAQ09569.1 GGMR01019223 MBY31842.1 GEZM01030681 JAV85429.1 GEZM01030680 JAV85430.1 ABLF02019274 ABLF02019277 ABLF02019278 ABLF02019280 ABLF02019281 ABLF02019283 GEZM01030679 JAV85431.1 GGMR01018115 MBY30734.1 GGMS01013823 MBY83026.1 GBGD01000837 JAC88052.1 ACPB03000713 KQ971318 EFA00393.2 EU597677 ACH86322.1 APGK01049566 APGK01049567 KB741156 KB632169 ENN73547.1 ERL89466.1 GEDC01014376 JAS22922.1 GEDC01021174 JAS16124.1 NEVH01019067 PNF23126.1 GEBQ01025115 GEBQ01018628 JAT14862.1 JAT21349.1 GAMC01020671 GAMC01020669 JAB85886.1 CH933809 EDW17819.1 GECZ01024254 JAS45515.1 KRG05778.1 KRG05779.1 GECZ01009466 JAS60303.1 CP012525 ALC44969.1 GBXI01012221 GBXI01009910 GBXI01003836 JAD02071.1 JAD04382.1 JAD10456.1 GDHF01021028 JAI31286.1 CM000159 KRK02018.1 CH954178 KQS44048.1 GDHF01022024 JAI30290.1 AE014296 BT033062 AAF49348.2 ACE82585.1 BT133104 AAN11709.1 AEX98024.1 AHN58118.1 CH902618 KPU78195.1 CH916366 EDV96998.1 BT099677 ACV44463.1 EDW94730.1 KRK02019.1 CM002912 KMZ00221.1 EDV51975.2 KQS44049.1 EDV39743.1 KPU78196.1 CM000363 EDX10844.1 CH480815 EDW41872.1 BT133440 AAN11707.1 AAN11708.1 AFH78572.1 BT099633 ACU64822.1 KMZ00220.1 KMZ00222.1 AJ243264 CAB53640.1 OUUW01000012 SPP87310.1 CH379069 EDY73688.2 CH963847 EDW73465.2 GAKP01020255 GAKP01020254 JAC38698.1 GFDF01007341 JAV06743.1 GFDF01007342 JAV06742.1 JXJN01023976 CCAG010006264 CH479214 EDW33444.1 JXJN01025645 GFDL01014880 JAV20165.1 KQ763581 OAD54951.1 DS232081 EDS34236.1 UFQS01002456 UFQT01002456 SSX14090.1 SSX33507.1 JXUM01047773 JXUM01047774 KQ561532 KXJ78248.1 ADMH02000413 ETN66607.1 GFDL01015025 JAV20020.1 CH477700 EAT37089.1 GFDL01015001 JAV20044.1 GFDL01014923 JAV20122.1 GGFJ01002097 MBW51238.1 GGFJ01002098 MBW51239.1 AJVK01033670 AJVK01033671 AJVK01033672 GGFM01000710 MBW21461.1
PZC72266.1 KQ459605 KPI92042.1 AGBW02013269 OWR43759.1 KQ460953 KPJ10306.1 KQ459603 KPI92908.1 KQ461194 KPJ06924.1 ODYU01003081 SOQ41391.1 NWSH01000927 PCG73520.1 BABH01005368 BABH01005369 BABH01005370 GFXV01003570 MBW15375.1 KC713794 AHH29252.1 GDHC01001187 JAQ17442.1 GDHC01009060 JAQ09569.1 GGMR01019223 MBY31842.1 GEZM01030681 JAV85429.1 GEZM01030680 JAV85430.1 ABLF02019274 ABLF02019277 ABLF02019278 ABLF02019280 ABLF02019281 ABLF02019283 GEZM01030679 JAV85431.1 GGMR01018115 MBY30734.1 GGMS01013823 MBY83026.1 GBGD01000837 JAC88052.1 ACPB03000713 KQ971318 EFA00393.2 EU597677 ACH86322.1 APGK01049566 APGK01049567 KB741156 KB632169 ENN73547.1 ERL89466.1 GEDC01014376 JAS22922.1 GEDC01021174 JAS16124.1 NEVH01019067 PNF23126.1 GEBQ01025115 GEBQ01018628 JAT14862.1 JAT21349.1 GAMC01020671 GAMC01020669 JAB85886.1 CH933809 EDW17819.1 GECZ01024254 JAS45515.1 KRG05778.1 KRG05779.1 GECZ01009466 JAS60303.1 CP012525 ALC44969.1 GBXI01012221 GBXI01009910 GBXI01003836 JAD02071.1 JAD04382.1 JAD10456.1 GDHF01021028 JAI31286.1 CM000159 KRK02018.1 CH954178 KQS44048.1 GDHF01022024 JAI30290.1 AE014296 BT033062 AAF49348.2 ACE82585.1 BT133104 AAN11709.1 AEX98024.1 AHN58118.1 CH902618 KPU78195.1 CH916366 EDV96998.1 BT099677 ACV44463.1 EDW94730.1 KRK02019.1 CM002912 KMZ00221.1 EDV51975.2 KQS44049.1 EDV39743.1 KPU78196.1 CM000363 EDX10844.1 CH480815 EDW41872.1 BT133440 AAN11707.1 AAN11708.1 AFH78572.1 BT099633 ACU64822.1 KMZ00220.1 KMZ00222.1 AJ243264 CAB53640.1 OUUW01000012 SPP87310.1 CH379069 EDY73688.2 CH963847 EDW73465.2 GAKP01020255 GAKP01020254 JAC38698.1 GFDF01007341 JAV06743.1 GFDF01007342 JAV06742.1 JXJN01023976 CCAG010006264 CH479214 EDW33444.1 JXJN01025645 GFDL01014880 JAV20165.1 KQ763581 OAD54951.1 DS232081 EDS34236.1 UFQS01002456 UFQT01002456 SSX14090.1 SSX33507.1 JXUM01047773 JXUM01047774 KQ561532 KXJ78248.1 ADMH02000413 ETN66607.1 GFDL01015025 JAV20020.1 CH477700 EAT37089.1 GFDL01015001 JAV20044.1 GFDL01014923 JAV20122.1 GGFJ01002097 MBW51238.1 GGFJ01002098 MBW51239.1 AJVK01033670 AJVK01033671 AJVK01033672 GGFM01000710 MBW21461.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000005204
UP000007819
+ More
UP000015103 UP000007266 UP000019118 UP000030742 UP000235965 UP000079169 UP000009192 UP000092553 UP000002282 UP000192221 UP000008711 UP000000803 UP000007801 UP000001070 UP000000304 UP000001292 UP000268350 UP000078200 UP000001819 UP000007798 UP000092443 UP000092460 UP000092444 UP000095301 UP000008744 UP000091820 UP000095300 UP000002320 UP000069940 UP000249989 UP000000673 UP000069272 UP000008820 UP000075901 UP000075902 UP000092462
UP000015103 UP000007266 UP000019118 UP000030742 UP000235965 UP000079169 UP000009192 UP000092553 UP000002282 UP000192221 UP000008711 UP000000803 UP000007801 UP000001070 UP000000304 UP000001292 UP000268350 UP000078200 UP000001819 UP000007798 UP000092443 UP000092460 UP000092444 UP000095301 UP000008744 UP000091820 UP000095300 UP000002320 UP000069940 UP000249989 UP000000673 UP000069272 UP000008820 UP000075901 UP000075902 UP000092462
Pfam
PF00209 SNF
SUPFAM
SSF161070
SSF161070
ProteinModelPortal
A0A2H1WUH2
A0A2A4JU52
A0A2A4JUW4
A0A2W1BHG8
A0A194PFI0
A0A212EQK9
+ More
A0A194R384 A0A194PIN1 A0A194QNJ1 A0A2H1VKR8 A0A2A4JQ39 H9JLX6 A0A2H8TMM4 X2D4J3 A0A146MEM0 A0A146LQK9 A0A2S2PQU7 A0A1Y1MI14 A0A1Y1ML21 J9KES3 A0A1Y1MIE3 A0A2S2PMX6 A0A2S2QZ54 A0A069DWH7 T1HU77 D6WED3 B6CY69 N6U4N6 A0A1B6DBA5 A0A1B6CRS2 A0A2J7Q3G4 A0A3Q0IT57 A0A1B6LCC7 W8AYE8 B4KZ22 A0A1B6F5R4 A0A0Q9XPD1 A0A1B6GDB9 A0A0M3QWZ4 A0A0A1WZX2 A0A0K8UXH4 A0A0R1E314 A0A1W4W4W9 A0A0Q5UJS4 A0A0K8UVD8 Q9VVG3 Q8IQQ6 A0A0P8YGX5 B4J247 C7LAC7 A0A1W4W4B2 B4PJW1 A0A0J9UN26 B3NHW0 B3M6C3 B4QNU2 B4HKR7 Q8IQQ7 C7LA83 A0A0J9RYC3 Q9U975 A0A3B0KG05 A0A1A9VYS9 B5DQ44 B4MMQ9 A0A034VAZ8 A0A1L8DJV5 A0A1L8DJV8 A0A1A9Y1H6 A0A1B0C1B4 A0A1B0FAG9 A0A1I8MQX4 B4H6L6 A0A1B0C4T3 A0A1A9WEB0 A0A1I8PP83 A0A1Q3EXX7 A0A310SK53 B0WT87 A0A336MYS6 A0A1S4FRX2 A0A1I8PP26 A0A182G8F8 W5JQB4 A0A1Q3EXK4 A0A182FEN9 Q16RP7 A0A1Q3EXK9 A0A1Q3EXU0 A0A182T2I6 A0A2M4BEJ5 A0A182THX6 A0A2M4BDW0 A0A1B0EY28 A0A2M3YYZ3
A0A194R384 A0A194PIN1 A0A194QNJ1 A0A2H1VKR8 A0A2A4JQ39 H9JLX6 A0A2H8TMM4 X2D4J3 A0A146MEM0 A0A146LQK9 A0A2S2PQU7 A0A1Y1MI14 A0A1Y1ML21 J9KES3 A0A1Y1MIE3 A0A2S2PMX6 A0A2S2QZ54 A0A069DWH7 T1HU77 D6WED3 B6CY69 N6U4N6 A0A1B6DBA5 A0A1B6CRS2 A0A2J7Q3G4 A0A3Q0IT57 A0A1B6LCC7 W8AYE8 B4KZ22 A0A1B6F5R4 A0A0Q9XPD1 A0A1B6GDB9 A0A0M3QWZ4 A0A0A1WZX2 A0A0K8UXH4 A0A0R1E314 A0A1W4W4W9 A0A0Q5UJS4 A0A0K8UVD8 Q9VVG3 Q8IQQ6 A0A0P8YGX5 B4J247 C7LAC7 A0A1W4W4B2 B4PJW1 A0A0J9UN26 B3NHW0 B3M6C3 B4QNU2 B4HKR7 Q8IQQ7 C7LA83 A0A0J9RYC3 Q9U975 A0A3B0KG05 A0A1A9VYS9 B5DQ44 B4MMQ9 A0A034VAZ8 A0A1L8DJV5 A0A1L8DJV8 A0A1A9Y1H6 A0A1B0C1B4 A0A1B0FAG9 A0A1I8MQX4 B4H6L6 A0A1B0C4T3 A0A1A9WEB0 A0A1I8PP83 A0A1Q3EXX7 A0A310SK53 B0WT87 A0A336MYS6 A0A1S4FRX2 A0A1I8PP26 A0A182G8F8 W5JQB4 A0A1Q3EXK4 A0A182FEN9 Q16RP7 A0A1Q3EXK9 A0A1Q3EXU0 A0A182T2I6 A0A2M4BEJ5 A0A182THX6 A0A2M4BDW0 A0A1B0EY28 A0A2M3YYZ3
PDB
4XPH
E-value=1.56033e-06,
Score=126
Ontologies
GO
PANTHER
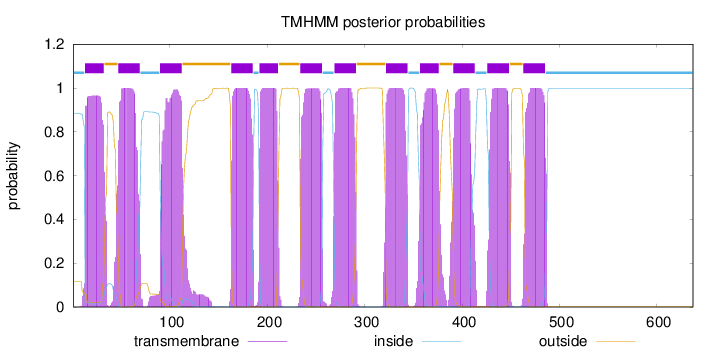
Topology
Length:
637
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
267.01281
Exp number, first 60 AAs:
35.90045
Total prob of N-in:
0.88442
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 32
outside
33 - 46
TMhelix
47 - 69
inside
70 - 89
TMhelix
90 - 112
outside
113 - 162
TMhelix
163 - 185
inside
186 - 191
TMhelix
192 - 211
outside
212 - 233
TMhelix
234 - 256
inside
257 - 268
TMhelix
269 - 291
outside
292 - 321
TMhelix
322 - 344
inside
345 - 356
TMhelix
357 - 376
outside
377 - 390
TMhelix
391 - 413
inside
414 - 425
TMhelix
426 - 448
outside
449 - 462
TMhelix
463 - 485
inside
486 - 637
Population Genetic Test Statistics
Pi
15.558396
Theta
17.547306
Tajima's D
-1.219275
CLR
0.095821
CSRT
0.0990450477476126
Interpretation
Uncertain
