Gene
KWMTBOMO00554
Pre Gene Modal
BGIBMGA012339
Annotation
PREDICTED:_E3_ubiquitin-protein_ligase_highwire_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.817 PlasmaMembrane Reliability : 1.177
Sequence
CDS
ATGAACATGTCGACCGAACGACTAGACATGATTCCCGGAATTGATCACAATGTGATGAAAGAGAACGCTTTGTTCCTCCTCGATTTGGCTCCACTCACTGATTCAGAACCTCTAAGCGTGTCTCCATGGAGCGAGAACTTGAGTCGAAGCCCCCCGACTTCGCCGGGGAGCGTATGGCAACCGGCGCCTCCCTTCCAATGCCTACAAGCACTCGGCGCTGAGCCTAAAGAGAACGACAACAACGACGCCGCCAAGTATCACTCGCTAGGACGTCCGCAACCCCCTGCCATCGCTCCTACAGTGTCGAATTTCGCCGCTCCGGAGGCCAGCCTTGGTTTCCAGCCCCGAGTCCACCGTTCTGTGTCTATGGGACAAGCCGGAGGAGACCTCGCGGCAGCCGCCCGACCTCTGCTCACTCGCAAGTTGCGTAGCGATTCGCATGACACCGCCACCGGTAGTGGATTGTCACTGCTCTCGCAACCGAGCAAGGCGCTGAAGGGTCTACTCAGCTGCGGCGATTGGAGTGCCACGGACTGTCCTTTGTCCGCATTTGAAGCCCCTCGACTAGACTCCAACATTATAATGAAAAGGCCGGTGCTGAGCTTCGTGCTCGCAAAACGCGATTTGGCCGCTCACAGAGAGAAAATGGACGCGGCCGTCAGGATTAACATTGTCAGACAATACGCGTTTGAGGCCCTCAATTGGCTGCTTCGTTCAGCGACACAGCCAATCTGCGCCCACGATGTTATGTGGTGGTTCTGTACAGCTTTGGAAAAGTTTGCCAGGATCGTACCACCACCGCACATTGTTGAAGATAATAAGGAGAATGCAGTAGCAGAGCCGATACGCACGGTCCCGCCAACCGCTTCGACGTCCGCCATCTGCCCCGGCGGCCGAGCAGCCCGCGGCGCCCGAGCTGCTTTCCACTCGTTCCTGGGCTCCGTCAGTACTTTAGCACCATCGCTACCACACGCCAGTGCCGCGAGTCTGCAGGCCATACGATGCTGGGCGCTCCACTACTCCAGCCACGATCGGGCTTTCTTGCACAGATCGCAAGTGTTCAGTGTAATCAGCAAGATATTATCTCGTTCTGAGGAAGGCGTCCACGACGACGGCCCACATTGTGCCATACACGACAGCTTCCATAGCTATTTGAAGGAGAATCATGTATGGAGCTCTTCCGACGTGACTGGCTGGTGCGATATTACGGTATCTTCACGTCAAGGCATGGCCGGCGCCTTAACCGACGGCAGCACGGAGACGTTCTGGGAATCAGGCGACGAAGACAGGAACAAAGCAAAGTGGATACAAATCTCTTACCCGGGAGGCTCGCACGATGATCGCCCGCACATTGTGTGCGTCCACATCGATAACACAAGGGACACTGTGAACAAGACGCTGCTGATGACGTTCCTGTACAGCTCAGGATCCACTGAGTTGATCCACATGCAAGACATAGAAGTCGATCCGAAGGTCGCTACTTGGCTCTGCTATACTTTACCACGTGTGTCTACCACGTCGCTGCGCGTTCGATGCGAGCTCCGCGGTCCCGAAGCGGCGGTTCGAATCCGCCAGGTCCGGGTCCTGAGCGCGCCGACGCTCCTCCATTCAGCCGCCGTGACAGCGCCGCAAGTGTTGCACGGACTCTGCGAAACGGACACCCTGCGAGTTTTCCGCTTGTTGACTTCTCAGGTGTTCGGGAAGCTCTTGGAATGGGAGCAGTCTAGCGACGCGAGCGCCGAAGCCGCGGTAGCAATCGACGATGGAGTTGCCAACGACGACAGCGATCTCAGAGAGCACGTTGTTGGCATCCTATTCGCTGGACACAAACTTACATCTCTGCAACGACAAGTGATGTCTCATATAGTAAGTGCGATCGGATGTGAGACTGCCCGCGTGCGCGATGATTGGGAGACAACTCTGCTTTGTGTCGAACCCACGGATAGTGAATGGGAGGAGTCGCCAGCCGCCAAACACGTGGGGTCGCAGCAGGACAACTACTGCTTTGAAATGCTCTCGCTGTTATTGGCACTCAGCGGCAGTGCAGTGGGTCGAGTACATCTTGCTCAGCGCACGGAACTACTATCTGACTTGTTATCTCTGCTTCACACTGGTAGCGAACGCGTACAAAGACAGGTAATATCGCTCTTACGTCGATTGGTCATAGAGCTACCGCCCCAGAAGATGTTAGCGGCCGTCAATTATGGTTCCGATCCGATTAGCAGGGTAACTTTACTCGACCATCTGGTTTGCTATTTGGGCAAAGCGATCACAGTTCAAATGAAGGTGAAAGGTTCAAGTGGATCGGCCCCAGCTACCGTTATGATGGGGGGCAGTATGGTGTCCTTGTCCGCGAGTAATTGGTTCATGAGAGGCGAGACTACCAAAAAACACGCACATTTGGTAGCTAAACTTCTATCTGACATGGCAGAGGATAAATTGGCTGGTTCATGGGGCCTAGAGACACGTACCCAATTAGCCAACTACGTATGCGCCATATCGCAAGTATCCGAAAAAGAGAGACAGCCGGCGCTTTGCATCGCGTCCCCCACTATCTGGATGGCGCTAGCTTCCCTGTGCGTGTGTGAACAATCACATCTAGATCTCGTCAACAGTTCTGGAGACGGGCGCGGAGCTCGTGGTCGAGACTCTTCCTCCGAGACTCGTCCTCTGTGCGCCAACCACGACGACGGCGCCACAGCGGCGGTCATAGACTGCCGCAATTGTGGACCGCTCTGTGCTGAATGCGATCGATTCTTGCACCTAAACAGGACCGCCAGGACTCACCACAGACAGATCTGCAAGGAGGAGGAGAGCGCTATACGCATCGACATACACGAAGGCTGTGGACGCGCGAAACTGTTTTGGCTATTGCTCCTGGTCGACAGGCGTACGCTGAAAGCCTTAGCCGAGTTCAGAGGAATGGAAGGTGGAGTCTGTGAAGCCGGACCATCATCTGAGGGACCGGCCGTGGCTTCGGGACCGGTGGCTCCCTCTGGAACGTGTCGCTTTTGCGGGGCCAGAGAATCTACCGGCATACTCACCATCGGAAATATCTGCGCCGATCAACAATGTCAGGACCATGGTCGGGAAGCATGTAATGTCATCCTCGCTTGCGGTCACCTGTGCGGTGGAGTTCGGGGAGAGCAAACCTGTCTGCCGTGCCTACATGGGTGCGCGGGCGTCTCAGCTGGTGAGGCAGCGCCACTGAGACAGGACGCGGACGATATGTGCATGATTTGCTTCACTGACCCCTTGCAGGCCGCCCCTGCGATACAGCTAACTTGCGGGCATGTGTTCCATCTGCACTGCTGCAGGAAGGTGCTCGTCAATAAGTGGAACGGTCCCAGGATTAGTTTCTCGTTCTCGCAGTGCCCAATTTGCAAGGAGGACATAAACCACTGGACCCTTGAAGAGTTGTTAACACCTATCAGGCAACTGCGTGATGAAGTGAAACGCAAGGCACTAATGAGACTCGAGTACGAGGGCCTGTCTGCTCCGGGCGGGACCAAAGGACGCAATGTTTCCGATCCGGCTTCCTACGCCATGGACAGATATGCTTATTACGTCTGTCATAAATGTGGCAAGGCCTATTTCGGCGGGCTGGCTCGTTGTGAGGCCGAGTCGAATGGCTGGTGGGAGCCGGCTGAGCTCGTGTGCGGCGCTTGCAGTGACGTCACCGGCGCTAGGACTTGCCCGAAACACGGAGCCGACTTTCTCGAGTACAAATGCCGGTATTGCTGTTCGGTTGCAGTGTTCTTCTGCTTCGGCACGTCGCACTTCTGCAACGCCTGCCACGACGACTTCCAGCGCGTCACACACATACCGCGACATCTGTTGCCTCAATGTCCAGCAGGTCCTAAAGGTGAACAGCTGCCGGGTACATCGGACGAGTGTCCTCTGCACGTGCAACATCCGGCGACCGGCGAGGAGTTCGCGCTCGGCTGCGGCGTGTGTCGGCACGCGCAAGGCTTCTAA
Protein
MNMSTERLDMIPGIDHNVMKENALFLLDLAPLTDSEPLSVSPWSENLSRSPPTSPGSVWQPAPPFQCLQALGAEPKENDNNDAAKYHSLGRPQPPAIAPTVSNFAAPEASLGFQPRVHRSVSMGQAGGDLAAAARPLLTRKLRSDSHDTATGSGLSLLSQPSKALKGLLSCGDWSATDCPLSAFEAPRLDSNIIMKRPVLSFVLAKRDLAAHREKMDAAVRINIVRQYAFEALNWLLRSATQPICAHDVMWWFCTALEKFARIVPPPHIVEDNKENAVAEPIRTVPPTASTSAICPGGRAARGARAAFHSFLGSVSTLAPSLPHASAASLQAIRCWALHYSSHDRAFLHRSQVFSVISKILSRSEEGVHDDGPHCAIHDSFHSYLKENHVWSSSDVTGWCDITVSSRQGMAGALTDGSTETFWESGDEDRNKAKWIQISYPGGSHDDRPHIVCVHIDNTRDTVNKTLLMTFLYSSGSTELIHMQDIEVDPKVATWLCYTLPRVSTTSLRVRCELRGPEAAVRIRQVRVLSAPTLLHSAAVTAPQVLHGLCETDTLRVFRLLTSQVFGKLLEWEQSSDASAEAAVAIDDGVANDDSDLREHVVGILFAGHKLTSLQRQVMSHIVSAIGCETARVRDDWETTLLCVEPTDSEWEESPAAKHVGSQQDNYCFEMLSLLLALSGSAVGRVHLAQRTELLSDLLSLLHTGSERVQRQVISLLRRLVIELPPQKMLAAVNYGSDPISRVTLLDHLVCYLGKAITVQMKVKGSSGSAPATVMMGGSMVSLSASNWFMRGETTKKHAHLVAKLLSDMAEDKLAGSWGLETRTQLANYVCAISQVSEKERQPALCIASPTIWMALASLCVCEQSHLDLVNSSGDGRGARGRDSSSETRPLCANHDDGATAAVIDCRNCGPLCAECDRFLHLNRTARTHHRQICKEEESAIRIDIHEGCGRAKLFWLLLLVDRRTLKALAEFRGMEGGVCEAGPSSEGPAVASGPVAPSGTCRFCGARESTGILTIGNICADQQCQDHGREACNVILACGHLCGGVRGEQTCLPCLHGCAGVSAGEAAPLRQDADDMCMICFTDPLQAAPAIQLTCGHVFHLHCCRKVLVNKWNGPRISFSFSQCPICKEDINHWTLEELLTPIRQLRDEVKRKALMRLEYEGLSAPGGTKGRNVSDPASYAMDRYAYYVCHKCGKAYFGGLARCEAESNGWWEPAELVCGACSDVTGARTCPKHGADFLEYKCRYCCSVAVFFCFGTSHFCNACHDDFQRVTHIPRHLLPQCPAGPKGEQLPGTSDECPLHVQHPATGEEFALGCGVCRHAQGF
Summary
Uniprot
H9JS29
A0A212EQJ5
A0A194PG88
D6WED4
A0A1Y1KG51
A0A195AUE6
+ More
E9IE92 A0A195D8J1 A0A158P185 A0A195DKF3 A0A026VU65 A0A151JUT3 E2BTI1 A0A1W5LU04 A0A3L8E0J1 A0A0J7L799 A0A087ZTJ5 A0A0L7QZ34 E2A5D5 A0A0T6B1U8 A0A0M8ZZX1 E0VWH2 A0A154P904 F4WU14 A0A310SWY6 T1JCF5 T1HM01 A0A2R5LJS7 A0A293MV39 A0A1B6II52 A0A1B6IU54 A0A2L2YC57 A0A2L2YC52 A0A1L8HHE3 A0A287AXI4 W5Q4B1 A0A1L8H976 A0A287AAA1 F1RP77 A0A2U4A649 A0A2U4A663 A0A287BIQ1 A0A287B464 A0A2Y9SNJ7 A0A2U4A604 A0A2U4A656 A0A2U4A614 A0A2U4A607 A0A2Y9STE2 A0A2Y9SKU1 A0A2Y9SK55 A0A2Y9NID4 A0A2Y9NIC4 A0A1S3EKT5 A0A1S3EKS4 A0A341C6X8 A0A2U4A6V0 A0A2U4A613 A0A2U4A6V5 A0A2U4A609 A0A2Y9SRF9 A0A1Z5KUW7 G3WS84 A0A2U4A618 A0A2Y9NMX8 A0A341C6Y7 A0A341C4B0 A0A2Y9NF46 A0A2Y9NF51 A0A2Y9NMY4 A0A2Y9NTD6 A0A2Y9NTE2 A0A341C4D8 A0A341C4B6 A0A341C5B9 G3U9L7 A0A2Y9NCP6 A0A2Y9NIC9 A0A2Y9NCQ1 A0A341C5Q5 A0A341C5R0 E1BH40 J9PB77 A0A2I0MSA8 F6SH29 V4AJP0 G1T291 G3WS85 A0A2Y9HUF8 A0A1V4J4C6 A0A287D8T7 D4A2D3 F7FJ16 K7EUD0 A0A2Y9JFD8 A0A2U3WNM5 A0A1V4J490 A0A1V4J435 A0A1V4J4E8 K9IPM6
E9IE92 A0A195D8J1 A0A158P185 A0A195DKF3 A0A026VU65 A0A151JUT3 E2BTI1 A0A1W5LU04 A0A3L8E0J1 A0A0J7L799 A0A087ZTJ5 A0A0L7QZ34 E2A5D5 A0A0T6B1U8 A0A0M8ZZX1 E0VWH2 A0A154P904 F4WU14 A0A310SWY6 T1JCF5 T1HM01 A0A2R5LJS7 A0A293MV39 A0A1B6II52 A0A1B6IU54 A0A2L2YC57 A0A2L2YC52 A0A1L8HHE3 A0A287AXI4 W5Q4B1 A0A1L8H976 A0A287AAA1 F1RP77 A0A2U4A649 A0A2U4A663 A0A287BIQ1 A0A287B464 A0A2Y9SNJ7 A0A2U4A604 A0A2U4A656 A0A2U4A614 A0A2U4A607 A0A2Y9STE2 A0A2Y9SKU1 A0A2Y9SK55 A0A2Y9NID4 A0A2Y9NIC4 A0A1S3EKT5 A0A1S3EKS4 A0A341C6X8 A0A2U4A6V0 A0A2U4A613 A0A2U4A6V5 A0A2U4A609 A0A2Y9SRF9 A0A1Z5KUW7 G3WS84 A0A2U4A618 A0A2Y9NMX8 A0A341C6Y7 A0A341C4B0 A0A2Y9NF46 A0A2Y9NF51 A0A2Y9NMY4 A0A2Y9NTD6 A0A2Y9NTE2 A0A341C4D8 A0A341C4B6 A0A341C5B9 G3U9L7 A0A2Y9NCP6 A0A2Y9NIC9 A0A2Y9NCQ1 A0A341C5Q5 A0A341C5R0 E1BH40 J9PB77 A0A2I0MSA8 F6SH29 V4AJP0 G1T291 G3WS85 A0A2Y9HUF8 A0A1V4J4C6 A0A287D8T7 D4A2D3 F7FJ16 K7EUD0 A0A2Y9JFD8 A0A2U3WNM5 A0A1V4J490 A0A1V4J435 A0A1V4J4E8 K9IPM6
Pubmed
EMBL
BABH01017139
BABH01017140
BABH01017141
BABH01017142
BABH01017143
AGBW02013269
+ More
OWR43758.1 KQ459605 KPI92043.1 KQ971318 EFA00365.2 GEZM01089083 JAV57727.1 KQ976738 KYM75801.1 GL762567 EFZ21106.1 KQ976749 KYN08749.1 ADTU01005969 ADTU01005970 KQ980765 KYN13336.1 KK107921 EZA47185.1 KQ981732 KYN36598.1 GL450410 EFN81009.1 KX034171 ANO54001.1 QOIP01000001 RLU26230.1 LBMM01000422 KMQ98404.1 KQ414683 KOC63870.1 GL436921 EFN71357.1 LJIG01016186 KRT81356.1 KQ435797 KOX73506.1 DS235820 EEB17728.1 KQ434846 KZC08416.1 GL888345 EGI62371.1 KQ759842 OAD62616.1 JH432064 ACPB03010408 GGLE01005676 MBY09802.1 GFWV01019103 MAA43831.1 GECU01021116 JAS86590.1 GECU01017258 JAS90448.1 IAAA01014318 LAA05718.1 IAAA01014319 LAA05723.1 CM004468 OCT95497.1 AEMK02000077 AMGL01015229 AMGL01015230 AMGL01015231 CM004469 OCT92648.1 GFJQ02008202 JAV98767.1 AEFK01130985 AEFK01130986 AEFK01130987 AEFK01130988 AAEX03013255 AAEX03013256 AAEX03013257 AKCR02000003 PKK32562.1 KB201890 ESO93791.1 AAGW02020608 AAGW02020609 AAGW02020610 AAGW02020611 LSYS01009367 OPJ66989.1 AGTP01036522 AGTP01036523 AGTP01036524 AGTP01036525 AABR07019086 AABR07019087 AABR07019088 AABR07019089 AABR07019090 AABR07019091 AABR07019092 AABR07019093 AABR07019094 AABR07019095 ABGA01182614 ABGA01182615 ABGA01182616 ABGA01182617 ABGA01182618 ABGA01182619 ABGA01182620 OPJ66988.1 OPJ66986.1 OPJ66990.1 GABZ01003325 JAA50200.1
OWR43758.1 KQ459605 KPI92043.1 KQ971318 EFA00365.2 GEZM01089083 JAV57727.1 KQ976738 KYM75801.1 GL762567 EFZ21106.1 KQ976749 KYN08749.1 ADTU01005969 ADTU01005970 KQ980765 KYN13336.1 KK107921 EZA47185.1 KQ981732 KYN36598.1 GL450410 EFN81009.1 KX034171 ANO54001.1 QOIP01000001 RLU26230.1 LBMM01000422 KMQ98404.1 KQ414683 KOC63870.1 GL436921 EFN71357.1 LJIG01016186 KRT81356.1 KQ435797 KOX73506.1 DS235820 EEB17728.1 KQ434846 KZC08416.1 GL888345 EGI62371.1 KQ759842 OAD62616.1 JH432064 ACPB03010408 GGLE01005676 MBY09802.1 GFWV01019103 MAA43831.1 GECU01021116 JAS86590.1 GECU01017258 JAS90448.1 IAAA01014318 LAA05718.1 IAAA01014319 LAA05723.1 CM004468 OCT95497.1 AEMK02000077 AMGL01015229 AMGL01015230 AMGL01015231 CM004469 OCT92648.1 GFJQ02008202 JAV98767.1 AEFK01130985 AEFK01130986 AEFK01130987 AEFK01130988 AAEX03013255 AAEX03013256 AAEX03013257 AKCR02000003 PKK32562.1 KB201890 ESO93791.1 AAGW02020608 AAGW02020609 AAGW02020610 AAGW02020611 LSYS01009367 OPJ66989.1 AGTP01036522 AGTP01036523 AGTP01036524 AGTP01036525 AABR07019086 AABR07019087 AABR07019088 AABR07019089 AABR07019090 AABR07019091 AABR07019092 AABR07019093 AABR07019094 AABR07019095 ABGA01182614 ABGA01182615 ABGA01182616 ABGA01182617 ABGA01182618 ABGA01182619 ABGA01182620 OPJ66988.1 OPJ66986.1 OPJ66990.1 GABZ01003325 JAA50200.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000007266
UP000078540
UP000078542
+ More
UP000005205 UP000078492 UP000053097 UP000078541 UP000008237 UP000279307 UP000036403 UP000005203 UP000053825 UP000000311 UP000053105 UP000009046 UP000076502 UP000007755 UP000015103 UP000186698 UP000008227 UP000002356 UP000245320 UP000248484 UP000248483 UP000081671 UP000252040 UP000007648 UP000007646 UP000009136 UP000002254 UP000053872 UP000002281 UP000030746 UP000001811 UP000248481 UP000190648 UP000005215 UP000002494 UP000002280 UP000001595 UP000248482 UP000245340
UP000005205 UP000078492 UP000053097 UP000078541 UP000008237 UP000279307 UP000036403 UP000005203 UP000053825 UP000000311 UP000053105 UP000009046 UP000076502 UP000007755 UP000015103 UP000186698 UP000008227 UP000002356 UP000245320 UP000248484 UP000248483 UP000081671 UP000252040 UP000007648 UP000007646 UP000009136 UP000002254 UP000053872 UP000002281 UP000030746 UP000001811 UP000248481 UP000190648 UP000005215 UP000002494 UP000002280 UP000001595 UP000248482 UP000245340
Pfam
Interpro
IPR004939
APC_su10/DOC_dom
+ More
IPR001841 Znf_RING
IPR013083 Znf_RING/FYVE/PHD
IPR008979 Galactose-bd-like_sf
IPR003646 SH3-like_bac-type
IPR014756 Ig_E-set
IPR017868 Filamin/ABP280_repeat-like
IPR012983 PHR
IPR009091 RCC1/BLIP-II
IPR038648 PHR_sf
IPR000408 Reg_chr_condens
IPR013087 Znf_C2H2_type
IPR000315 Znf_B-box
IPR016024 ARM-type_fold
IPR001841 Znf_RING
IPR013083 Znf_RING/FYVE/PHD
IPR008979 Galactose-bd-like_sf
IPR003646 SH3-like_bac-type
IPR014756 Ig_E-set
IPR017868 Filamin/ABP280_repeat-like
IPR012983 PHR
IPR009091 RCC1/BLIP-II
IPR038648 PHR_sf
IPR000408 Reg_chr_condens
IPR013087 Znf_C2H2_type
IPR000315 Znf_B-box
IPR016024 ARM-type_fold
ProteinModelPortal
H9JS29
A0A212EQJ5
A0A194PG88
D6WED4
A0A1Y1KG51
A0A195AUE6
+ More
E9IE92 A0A195D8J1 A0A158P185 A0A195DKF3 A0A026VU65 A0A151JUT3 E2BTI1 A0A1W5LU04 A0A3L8E0J1 A0A0J7L799 A0A087ZTJ5 A0A0L7QZ34 E2A5D5 A0A0T6B1U8 A0A0M8ZZX1 E0VWH2 A0A154P904 F4WU14 A0A310SWY6 T1JCF5 T1HM01 A0A2R5LJS7 A0A293MV39 A0A1B6II52 A0A1B6IU54 A0A2L2YC57 A0A2L2YC52 A0A1L8HHE3 A0A287AXI4 W5Q4B1 A0A1L8H976 A0A287AAA1 F1RP77 A0A2U4A649 A0A2U4A663 A0A287BIQ1 A0A287B464 A0A2Y9SNJ7 A0A2U4A604 A0A2U4A656 A0A2U4A614 A0A2U4A607 A0A2Y9STE2 A0A2Y9SKU1 A0A2Y9SK55 A0A2Y9NID4 A0A2Y9NIC4 A0A1S3EKT5 A0A1S3EKS4 A0A341C6X8 A0A2U4A6V0 A0A2U4A613 A0A2U4A6V5 A0A2U4A609 A0A2Y9SRF9 A0A1Z5KUW7 G3WS84 A0A2U4A618 A0A2Y9NMX8 A0A341C6Y7 A0A341C4B0 A0A2Y9NF46 A0A2Y9NF51 A0A2Y9NMY4 A0A2Y9NTD6 A0A2Y9NTE2 A0A341C4D8 A0A341C4B6 A0A341C5B9 G3U9L7 A0A2Y9NCP6 A0A2Y9NIC9 A0A2Y9NCQ1 A0A341C5Q5 A0A341C5R0 E1BH40 J9PB77 A0A2I0MSA8 F6SH29 V4AJP0 G1T291 G3WS85 A0A2Y9HUF8 A0A1V4J4C6 A0A287D8T7 D4A2D3 F7FJ16 K7EUD0 A0A2Y9JFD8 A0A2U3WNM5 A0A1V4J490 A0A1V4J435 A0A1V4J4E8 K9IPM6
E9IE92 A0A195D8J1 A0A158P185 A0A195DKF3 A0A026VU65 A0A151JUT3 E2BTI1 A0A1W5LU04 A0A3L8E0J1 A0A0J7L799 A0A087ZTJ5 A0A0L7QZ34 E2A5D5 A0A0T6B1U8 A0A0M8ZZX1 E0VWH2 A0A154P904 F4WU14 A0A310SWY6 T1JCF5 T1HM01 A0A2R5LJS7 A0A293MV39 A0A1B6II52 A0A1B6IU54 A0A2L2YC57 A0A2L2YC52 A0A1L8HHE3 A0A287AXI4 W5Q4B1 A0A1L8H976 A0A287AAA1 F1RP77 A0A2U4A649 A0A2U4A663 A0A287BIQ1 A0A287B464 A0A2Y9SNJ7 A0A2U4A604 A0A2U4A656 A0A2U4A614 A0A2U4A607 A0A2Y9STE2 A0A2Y9SKU1 A0A2Y9SK55 A0A2Y9NID4 A0A2Y9NIC4 A0A1S3EKT5 A0A1S3EKS4 A0A341C6X8 A0A2U4A6V0 A0A2U4A613 A0A2U4A6V5 A0A2U4A609 A0A2Y9SRF9 A0A1Z5KUW7 G3WS84 A0A2U4A618 A0A2Y9NMX8 A0A341C6Y7 A0A341C4B0 A0A2Y9NF46 A0A2Y9NF51 A0A2Y9NMY4 A0A2Y9NTD6 A0A2Y9NTE2 A0A341C4D8 A0A341C4B6 A0A341C5B9 G3U9L7 A0A2Y9NCP6 A0A2Y9NIC9 A0A2Y9NCQ1 A0A341C5Q5 A0A341C5R0 E1BH40 J9PB77 A0A2I0MSA8 F6SH29 V4AJP0 G1T291 G3WS85 A0A2Y9HUF8 A0A1V4J4C6 A0A287D8T7 D4A2D3 F7FJ16 K7EUD0 A0A2Y9JFD8 A0A2U3WNM5 A0A1V4J490 A0A1V4J435 A0A1V4J4E8 K9IPM6
PDB
5O6C
E-value=6.03558e-108,
Score=1004
Ontologies
GO
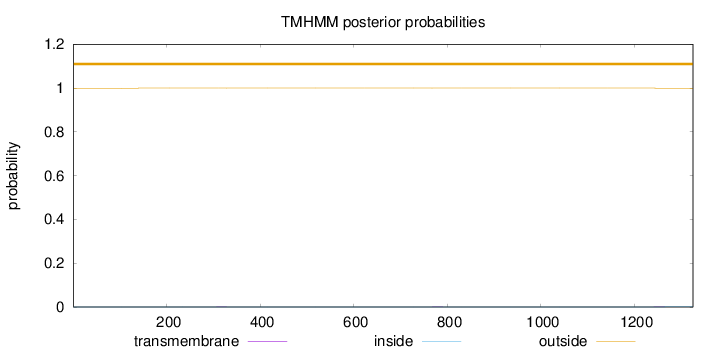
Topology
Length:
1325
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00040
outside
1 - 1325
Population Genetic Test Statistics
Pi
24.936591
Theta
22.208931
Tajima's D
0.177256
CLR
0.012824
CSRT
0.418379081045948
Interpretation
Uncertain
