Gene
KWMTBOMO00551
Pre Gene Modal
BGIBMGA012249
Annotation
Uncharacterized_protein_OBRU01_11868_[Operophtera_brumata]
Location in the cell
PlasmaMembrane Reliability : 3.855
Sequence
CDS
ATGCGTTGGACGACGTGGTGGCAGATTATTTTCGTGTTAAACACCTCCCTGGCACAGAGCAACACGGTAACCGTCTGCCAGGAGGATGATTGTAGCTGCGACTCATTCACGAGACTCATATGCAACTGCACCAGTGCAGACTCGGAGGTAACCCTAAGACCAGATGGCCCATACCGGGTACCATCCACCATCACAGCAATTTTTATCAACGGATGCGGACGAATATCATTCCTGTCGGATACCGTCCGAGATCTTATCAATCTACGGAACATCGAGATCAGAAATGTTCAACACGTTATCATCAATGAGAGAGCTTTGGCGTGGTTGCCGTTTCCGAGGGAAAATGAAATGAATCCAGGTCTCAGGATAACAATAGACAATTCAACGATTGATGAAATAGCGTCACACGCAATACAAGGGCGGTTTAACGACATCATAATCAGGAACAGCAAATTTGTTAATGCAAGACCTTTCGCGTTTTCTAGTCTTTTCGGTGTTAATAATGTAGAACTTACGAACAATGTGTACGAAAATATGGACATACAAGTCTTCAAAAAATTCTCAATGTTAAATTTCGTAATAAGCGGAGGTACCGTTAACGTTTTACCCAGTAGGTTTCTCTCTGACGTAGAAGTTACGAATCTTTTCCGCGTGGAAGGAGTTACCGCCAGATCCTTGTATAGTTTAACGTTCCTTGTTAGCTCACCAAAGAGGGTCCTTATCGAAAATAACAATATAGAAACAGTGGAAGGAGACGCCTTTCATATAGTCACAAGGGGACCCATAACTTTTAGAAATAATACTGTGACAAACTTGAAAAAGGGTGCCTTACTTGGATTCACAGTAGAATTGGAAGTAGCATCTGTGTCAGGAGCACAAGAATTATTGATTGACAATAATACGATAACGAATTTAAACCCATCATCGTTAATGTACAATCGCACTCGACTGAATCGGCGAGTGGACGGTTTAAACATAGACGTACCATGCAGTTGTGAACTAGCCGACGAATGGCGTAAAGCCCTTAAAGACGAAGGCGGAAATATTGCGTGTTGGTACGAACTAGAAAGATATTTTGTCTCTTTGCCGACTTACACCGACAGTAGGTGCGGTCCATTCAAGCAGAATTTCTGGATTTTTGTTGTTATTGGTATAGTATTAGCGTTAACTCTCGTAGCTATTGCGATATTTTTCATTGTAAGACATGAAAATGAAAAGAAAAAAAAGGTGCAGATAGTTATGCCAGATGGAAAGACTTACAGAGAGACTGAATTGCATATAGTAGTAGAAAGGGCCGAATTACTGACCACCGAATTATGA
Protein
MRWTTWWQIIFVLNTSLAQSNTVTVCQEDDCSCDSFTRLICNCTSADSEVTLRPDGPYRVPSTITAIFINGCGRISFLSDTVRDLINLRNIEIRNVQHVIINERALAWLPFPRENEMNPGLRITIDNSTIDEIASHAIQGRFNDIIIRNSKFVNARPFAFSSLFGVNNVELTNNVYENMDIQVFKKFSMLNFVISGGTVNVLPSRFLSDVEVTNLFRVEGVTARSLYSLTFLVSSPKRVLIENNNIETVEGDAFHIVTRGPITFRNNTVTNLKKGALLGFTVELEVASVSGAQELLIDNNTITNLNPSSLMYNRTRLNRRVDGLNIDVPCSCELADEWRKALKDEGGNIACWYELERYFVSLPTYTDSRCGPFKQNFWIFVVIGIVLALTLVAIAIFFIVRHENEKKKKVQIVMPDGKTYRETELHIVVERAELLTTEL
Summary
Uniprot
H9JRT9
A0A2H1WNC8
A0A2A4J878
A0A0L7LBQ4
A0A194QZA8
A0A194PFA3
+ More
A0A212EQI8 A0A1E1WMQ9 A0A069DZ28 A0A224XB66 N6SWR9 F4WU10 A0A195AU86 A0A158P115 U4UR85 A0A151JV29 A0A195D792 A0A1B6GG68 E0VWG8 A0A1Y1LBU5 D6WED6 A0A1B6K264 A0A1B6BZ64 A0A1B6MB45 A0A2J7QEF3 A0A1W4XG98 A0A1B6ELF9 A0A1B6M4L3 A0A0A9YL58 E2A5D7 T1HC93 A0A151WWZ4 A0A087ZTN1 A0A026VTS8 A0A3L8E0S1 A0A1B6JLU8 A0A195DLF7 E2BTH8 A0A067RFD2 A0A2S2R7L1 A0A0J7L412 A0A2P8Y1K9 E9IE89 X1WKL8 J9K8D4 A0A0P4VH97 A0A3Q0JKG2 B0W7P7 A0A1S4F6K9 Q17DR9 A0A1L8EC21 A0A1I8N9M2 A0A1I8P1W3 A0A1L8EBP8 A0A0L0CE25 A0A1L8EBN5 A0A1Q3FJE0 A0A034VQ99 A0A1S4EPT5 T1PEK7 W8C926 A0A1L8E8N3 A0A1L8E8S5 A0A1A9WVB3 A0A1W7R811 A0A023ES39 B4L252 B4PW96 B4H2J1 B4R4X7 B3NUW2 A0A1L8E7G3 A0A1A9Y9C3 B5DLG1
A0A212EQI8 A0A1E1WMQ9 A0A069DZ28 A0A224XB66 N6SWR9 F4WU10 A0A195AU86 A0A158P115 U4UR85 A0A151JV29 A0A195D792 A0A1B6GG68 E0VWG8 A0A1Y1LBU5 D6WED6 A0A1B6K264 A0A1B6BZ64 A0A1B6MB45 A0A2J7QEF3 A0A1W4XG98 A0A1B6ELF9 A0A1B6M4L3 A0A0A9YL58 E2A5D7 T1HC93 A0A151WWZ4 A0A087ZTN1 A0A026VTS8 A0A3L8E0S1 A0A1B6JLU8 A0A195DLF7 E2BTH8 A0A067RFD2 A0A2S2R7L1 A0A0J7L412 A0A2P8Y1K9 E9IE89 X1WKL8 J9K8D4 A0A0P4VH97 A0A3Q0JKG2 B0W7P7 A0A1S4F6K9 Q17DR9 A0A1L8EC21 A0A1I8N9M2 A0A1I8P1W3 A0A1L8EBP8 A0A0L0CE25 A0A1L8EBN5 A0A1Q3FJE0 A0A034VQ99 A0A1S4EPT5 T1PEK7 W8C926 A0A1L8E8N3 A0A1L8E8S5 A0A1A9WVB3 A0A1W7R811 A0A023ES39 B4L252 B4PW96 B4H2J1 B4R4X7 B3NUW2 A0A1L8E7G3 A0A1A9Y9C3 B5DLG1
Pubmed
EMBL
BABH01017129
ODYU01009814
SOQ54486.1
NWSH01002564
PCG67963.1
PCG67966.1
+ More
JTDY01001884 KOB72641.1 KQ460953 KPJ10310.1 KQ459605 KPI92046.1 AGBW02013269 OWR43756.1 GDQN01004559 GDQN01002764 JAT86495.1 JAT88290.1 GBGD01001365 JAC87524.1 GFTR01006779 JAW09647.1 APGK01053658 APGK01053659 KB741231 ENN72239.1 GL888345 EGI62367.1 KQ976738 KYM75798.1 ADTU01005953 ADTU01005954 KI206990 ERL95617.1 KQ981732 KYN36596.1 KQ976749 KYN08746.1 GECZ01008331 JAS61438.1 DS235820 EEB17724.1 GEZM01062337 GEZM01062336 GEZM01062335 GEZM01062334 JAV69810.1 KQ971318 EFA00395.1 GECU01002164 JAT05543.1 GEDC01030745 JAS06553.1 GEBQ01006827 JAT33150.1 NEVH01015312 PNF26951.1 GECZ01031010 JAS38759.1 GEBQ01009133 JAT30844.1 GBHO01013345 GBHO01013344 GDHC01008188 JAG30259.1 JAG30260.1 JAQ10441.1 GL436921 EFN71359.1 ACPB03010415 ACPB03010416 ACPB03010417 KQ982691 KYQ52185.1 KK107921 EZA47188.1 QOIP01000001 RLU26257.1 GECU01007498 JAT00209.1 KQ980765 KYN13334.1 GL450410 EFN81006.1 KK852542 KDR21728.1 GGMS01016710 MBY85913.1 LBMM01000728 KMQ97617.1 PYGN01001048 PSN38142.1 GL762567 EFZ21113.1 ABLF02009490 ABLF02010799 GDKW01003650 JAI52945.1 DS231855 EDS38191.1 CH477291 EAT44595.1 GFDG01002640 JAV16159.1 GFDG01002641 JAV16158.1 JRES01000509 KNC30500.1 GFDG01002639 JAV16160.1 GFDL01007304 JAV27741.1 GAKP01015250 GAKP01015249 JAC43703.1 KA646363 AFP60992.1 GAMC01002530 GAMC01002529 JAC04027.1 GFDG01003681 JAV15118.1 GFDG01003629 JAV15170.1 GEHC01000366 JAV47279.1 GAPW01002052 JAC11546.1 CH933810 EDW06792.1 CM000162 EDX01727.1 KRK06324.1 KRK06325.1 CH479204 EDW30558.1 CM000366 EDX17934.1 CH954180 EDV47060.1 GFDG01004156 JAV14643.1 CH379063 EDY72360.1
JTDY01001884 KOB72641.1 KQ460953 KPJ10310.1 KQ459605 KPI92046.1 AGBW02013269 OWR43756.1 GDQN01004559 GDQN01002764 JAT86495.1 JAT88290.1 GBGD01001365 JAC87524.1 GFTR01006779 JAW09647.1 APGK01053658 APGK01053659 KB741231 ENN72239.1 GL888345 EGI62367.1 KQ976738 KYM75798.1 ADTU01005953 ADTU01005954 KI206990 ERL95617.1 KQ981732 KYN36596.1 KQ976749 KYN08746.1 GECZ01008331 JAS61438.1 DS235820 EEB17724.1 GEZM01062337 GEZM01062336 GEZM01062335 GEZM01062334 JAV69810.1 KQ971318 EFA00395.1 GECU01002164 JAT05543.1 GEDC01030745 JAS06553.1 GEBQ01006827 JAT33150.1 NEVH01015312 PNF26951.1 GECZ01031010 JAS38759.1 GEBQ01009133 JAT30844.1 GBHO01013345 GBHO01013344 GDHC01008188 JAG30259.1 JAG30260.1 JAQ10441.1 GL436921 EFN71359.1 ACPB03010415 ACPB03010416 ACPB03010417 KQ982691 KYQ52185.1 KK107921 EZA47188.1 QOIP01000001 RLU26257.1 GECU01007498 JAT00209.1 KQ980765 KYN13334.1 GL450410 EFN81006.1 KK852542 KDR21728.1 GGMS01016710 MBY85913.1 LBMM01000728 KMQ97617.1 PYGN01001048 PSN38142.1 GL762567 EFZ21113.1 ABLF02009490 ABLF02010799 GDKW01003650 JAI52945.1 DS231855 EDS38191.1 CH477291 EAT44595.1 GFDG01002640 JAV16159.1 GFDG01002641 JAV16158.1 JRES01000509 KNC30500.1 GFDG01002639 JAV16160.1 GFDL01007304 JAV27741.1 GAKP01015250 GAKP01015249 JAC43703.1 KA646363 AFP60992.1 GAMC01002530 GAMC01002529 JAC04027.1 GFDG01003681 JAV15118.1 GFDG01003629 JAV15170.1 GEHC01000366 JAV47279.1 GAPW01002052 JAC11546.1 CH933810 EDW06792.1 CM000162 EDX01727.1 KRK06324.1 KRK06325.1 CH479204 EDW30558.1 CM000366 EDX17934.1 CH954180 EDV47060.1 GFDG01004156 JAV14643.1 CH379063 EDY72360.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000019118 UP000007755 UP000078540 UP000005205 UP000030742 UP000078541 UP000078542 UP000009046 UP000007266 UP000235965 UP000192223 UP000000311 UP000015103 UP000075809 UP000005203 UP000053097 UP000279307 UP000078492 UP000008237 UP000027135 UP000036403 UP000245037 UP000007819 UP000079169 UP000002320 UP000008820 UP000095301 UP000095300 UP000037069 UP000091820 UP000009192 UP000002282 UP000008744 UP000000304 UP000008711 UP000092443 UP000001819
UP000019118 UP000007755 UP000078540 UP000005205 UP000030742 UP000078541 UP000078542 UP000009046 UP000007266 UP000235965 UP000192223 UP000000311 UP000015103 UP000075809 UP000005203 UP000053097 UP000279307 UP000078492 UP000008237 UP000027135 UP000036403 UP000245037 UP000007819 UP000079169 UP000002320 UP000008820 UP000095301 UP000095300 UP000037069 UP000091820 UP000009192 UP000002282 UP000008744 UP000000304 UP000008711 UP000092443 UP000001819
PRIDE
Interpro
SUPFAM
SSF51126
SSF51126
Gene 3D
ProteinModelPortal
H9JRT9
A0A2H1WNC8
A0A2A4J878
A0A0L7LBQ4
A0A194QZA8
A0A194PFA3
+ More
A0A212EQI8 A0A1E1WMQ9 A0A069DZ28 A0A224XB66 N6SWR9 F4WU10 A0A195AU86 A0A158P115 U4UR85 A0A151JV29 A0A195D792 A0A1B6GG68 E0VWG8 A0A1Y1LBU5 D6WED6 A0A1B6K264 A0A1B6BZ64 A0A1B6MB45 A0A2J7QEF3 A0A1W4XG98 A0A1B6ELF9 A0A1B6M4L3 A0A0A9YL58 E2A5D7 T1HC93 A0A151WWZ4 A0A087ZTN1 A0A026VTS8 A0A3L8E0S1 A0A1B6JLU8 A0A195DLF7 E2BTH8 A0A067RFD2 A0A2S2R7L1 A0A0J7L412 A0A2P8Y1K9 E9IE89 X1WKL8 J9K8D4 A0A0P4VH97 A0A3Q0JKG2 B0W7P7 A0A1S4F6K9 Q17DR9 A0A1L8EC21 A0A1I8N9M2 A0A1I8P1W3 A0A1L8EBP8 A0A0L0CE25 A0A1L8EBN5 A0A1Q3FJE0 A0A034VQ99 A0A1S4EPT5 T1PEK7 W8C926 A0A1L8E8N3 A0A1L8E8S5 A0A1A9WVB3 A0A1W7R811 A0A023ES39 B4L252 B4PW96 B4H2J1 B4R4X7 B3NUW2 A0A1L8E7G3 A0A1A9Y9C3 B5DLG1
A0A212EQI8 A0A1E1WMQ9 A0A069DZ28 A0A224XB66 N6SWR9 F4WU10 A0A195AU86 A0A158P115 U4UR85 A0A151JV29 A0A195D792 A0A1B6GG68 E0VWG8 A0A1Y1LBU5 D6WED6 A0A1B6K264 A0A1B6BZ64 A0A1B6MB45 A0A2J7QEF3 A0A1W4XG98 A0A1B6ELF9 A0A1B6M4L3 A0A0A9YL58 E2A5D7 T1HC93 A0A151WWZ4 A0A087ZTN1 A0A026VTS8 A0A3L8E0S1 A0A1B6JLU8 A0A195DLF7 E2BTH8 A0A067RFD2 A0A2S2R7L1 A0A0J7L412 A0A2P8Y1K9 E9IE89 X1WKL8 J9K8D4 A0A0P4VH97 A0A3Q0JKG2 B0W7P7 A0A1S4F6K9 Q17DR9 A0A1L8EC21 A0A1I8N9M2 A0A1I8P1W3 A0A1L8EBP8 A0A0L0CE25 A0A1L8EBN5 A0A1Q3FJE0 A0A034VQ99 A0A1S4EPT5 T1PEK7 W8C926 A0A1L8E8N3 A0A1L8E8S5 A0A1A9WVB3 A0A1W7R811 A0A023ES39 B4L252 B4PW96 B4H2J1 B4R4X7 B3NUW2 A0A1L8E7G3 A0A1A9Y9C3 B5DLG1
Ontologies
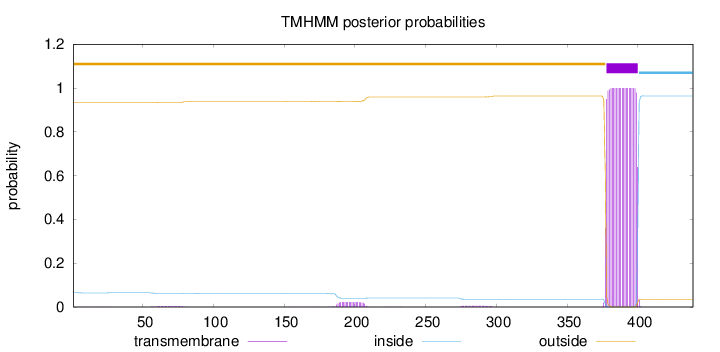
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.998029
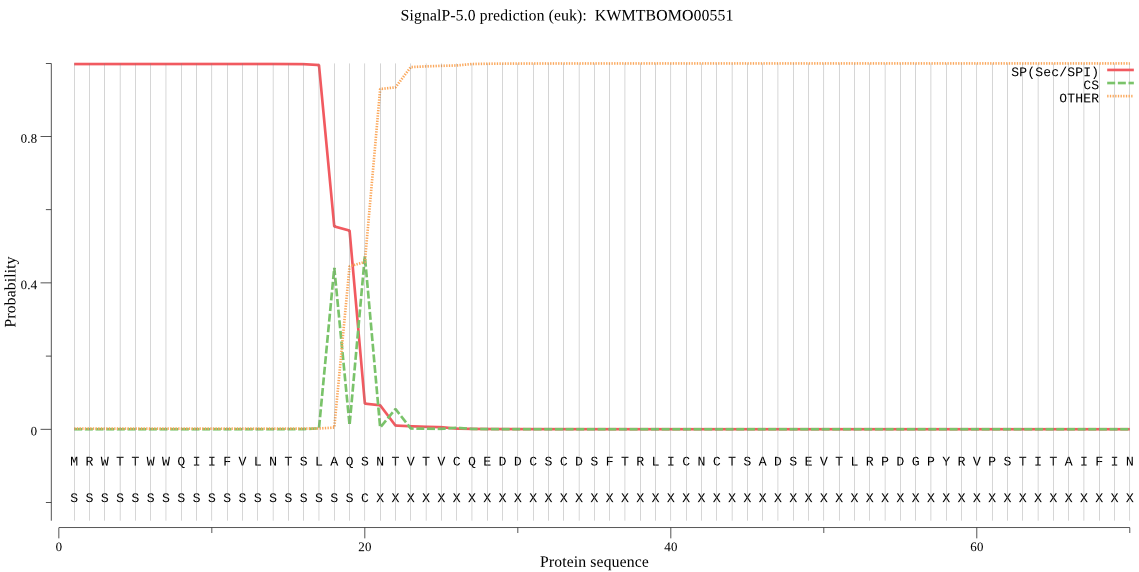
Length:
439
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.64504
Exp number, first 60 AAs:
0.02821
Total prob of N-in:
0.06483
outside
1 - 377
TMhelix
378 - 400
inside
401 - 439
Population Genetic Test Statistics
Pi
6.213922
Theta
19.2808
Tajima's D
-0.804083
CLR
0.134225
CSRT
0.176341182940853
Interpretation
Uncertain
