Gene
KWMTBOMO00548
Pre Gene Modal
BGIBMGA012252
Annotation
PREDICTED:_kalirin_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.71 Nuclear Reliability : 1.656
Sequence
CDS
ATGGAGGCAGGAGCAAGGGCTCTCGACGTCCTCCCGCTTCTGCAGGAGCGGGTGGCGTCTCTCACTGGTGGTCGGGATCGCCGGGGAAGTCCTATCATCTGGTTTCCGGCCAACTCAAGGAGGGATCGTGTCGCTCCAGATGACTATAGACGTTTGATACATTACTTGATCAGTATACCTGGTGAATCTGTGAAAAGTCTCGGATTCACAATATTAATTGACATGAGAGGTTCGGCGTGGGCTGCTATCAAGCCAATCCTAAAGGTTCTTCAAGAACACTTCCCATCATCCGTCCATCAGGCACTGGTAGTTAAACCTGACAATTTTTGGCAGAAACAACGTACAACTATTGGAGCACATAAATACAAATTTGAGACAACAATGATTAGTTTAGAAGCACTTCCTAAAGTTATAGATAGTTCACAATTGACTCCAGATCTAGAGGGTACATTGCAGTATGATCACCCACAGTGGATTGATCTAAGATTGGCATTAGAAGAATTGATGTGGCAGGCCGGTGAGCTTCTAGATCGATTAGATGATTTACAAGAGGATGTAGCCAGAGCAGACTTTGCTGATGATGTGACGGGTGCAAGGAGAGCCATTGATGCACATGCAGACATTAGTAAACGACTTGCCAAAGTTCCTGTTGATGAATTAGAAGCACAAGGTGAGAGGGTGTTACAACGATTAGAAGTGGCGGAGGCCGCGTGTTCCGAAGCTAGCGGTAGTGGCGCCGAAGGTTCTGCTTTCCACTGCGGATCGCCGGGCGCCGTGCGAGCTCAACTTTCGGCTGTGCGGTCCGCGCATGCGCATGCCCATAAACTTTGGCAACACAAAAAAATGCAGCTAGATCAGTGTTTTCAGCTCAGATTATTTGAACAAGACTGCGAGAAGATGTTGGAATGGATCGTGAACCACCGTACGGCCTTCTTAGCCACGTACGTTGAGATCGGACGGTCATGTGCGGCCGCAAAACGTTTACAGGAAGAACACGCTAGATTTGCAGCGGCGTGTACTAGCGGCGGAAGGCCGAGTGTCGCACGTGTGACGACAGCCGCGCGACGTCTCGCTGACAAGAGACATTACGCTGAAAGTCAGATATCAGCATTAGCACAAAGACTCGAAAGAGCTTACAAACAGTTGGCAGCCGGTCTTGAAGAACGTTCTGCAGTGCTGTCACTGTCGGTGGTATTTCATCATAAAGCTGAGGCGTACGCTTCAGCGGTGGGAGGGTGGGGAGCGCAGTGTGCCGTCGCTCTACAACTAGCAACGGGTCAAGGAGGAGCGCCTTGTAACGATCCGAGGGCATTAGAACAACACGCCCAGAGACATCGCCAACTTTACGAACACATGTGTCAGGCCTACACTGAGGTTCATTCAACAAGTAAAAAACTTTTATATCAGTTGGATCATTTAGTTCAAGTATGTCATCAAACGGATCCCACGGATATGCAGAGTGAAGGCACAGTGAGTAGTGCGGGTAGCAGTGGTGGCGATCCTGCAGCCGATTACAGTTCGGGAGCAAACCACGTGCTGGCCGTAATACATCAAATTCTTGGCCATCACAGGGCTTTGGAAGCTAGATATCACCAAGCGAGACTTAAATTGCATCAAAGATTAGCTTTGTTGCTCTATAAAGAAGACTGTAGACAGGTATTAGACTGGTTGGCAAACCATGGTGAAGTATTTTTGCAGAAAAATACTGGTATAGGAAGAAATCTCCATAAAGCTAGGGTTTATCAGAAATCTCATGAACATTTTGAAAATGTTGCACAAAATACTTACACAAACGCGGAGAAGCTGCTGGGAGCTGCTGAGGAGTTAGCCCGTTCGGGTGAATGTGATCCAGAGGAAGTCCTTGGAGTAGCGCGGGAACTCGAAGCTCATGTAGCTGCCTTCGCGGCCAGGGTCGATAGAAGAAGAAGACGACTCGACCTGGCCGTACTTCTGTATACACATGAGAAAGAGCTGCTGCAATGGTTGGATACACTGAGAAGCGGAGGCGGTGTATGCGCTACAGACGACACTCCCGAAGCGGCACGAAGAGCTTTAGAACAATGCGCCCAGCATCGAGCAGCTAGCATGGACGCATGCGCTGCAACTATCGCGCAGGGCGAAGCATTAATGCAAGACTTAAGATCTTCTGGCGATTCGGATGCAACCAGCACCGCTGCCGTGGAATCCACTTTAGAGCGATTGCGAGGAACACGCAGTGCACTCGAGGAGCTGTGGTCCGATCGCGAAAGGAAACTGGAGCTCACATTACAGCTTAGACATTTTGAAAGGGACGCTCTGGAGGTTTCGAGCCGACTAGAGTTGTGGGGTGACGAATTGCAACGCACCGAACCGCCTCGTGATCCCCAGCAAGCGGAGCAGGCCCTTGCAGCACACAACGAAAGTGTTGCTAGAATGCAACACGCTACTTTTCAGGTGGTACAGCAGGGTCAAGAATTGGCAGCAGCTATTGCAGACGCTTCAGATGTAATGGCATCTAGTGGTTCTGACGGGACTGAAACTGCGCCTCTTGATGCACAAGGTCGTGTGCAGCTGCTGCTTGAGTTCCTTCACGACAGGCAGCTTGATTTGGAAGAATTGGCTGAAGAAAGACGTGCCAGATTCGAACAGTGCGTGCAACTTGGACAATTTCAAAAAGACGCAGCTCAGGTGGTCAGTTGGATACGTAACGGAGAGGCTATGTTGGCTGCGTCGTTCTCCATTCCTGGAACTCTCTCTGAGGCCGAGCAGCTTAAAAGGGAGCACGACCAATTTCAAGTGGCAGTGGAGAAAACACACGCCAGTGCTGTTCAAGTAAAGTACAGGGCTGATGCGTTGAGGGCTGCCAATCACTACGATCCCCATACCATCAGAGAAATATCAGAGGAGGTAACTGAGCGCTGGCAGCGATTAGTGACCTGCGCCGAGGAGCGTCACAAGCTCGTGACGGCCTCACTCAATTTCTACAAGACAGCCGAACAGGTGTGTTCGGTGCTCGACTCATTGGAACGTGAATACCGCAGAGATGAAGACTGGTGTGGATCTGCAGCCGCTCCATCAACTGACGAGCCGACTTTAGCTCAGTCTATTGACAAAGCCGCACAGGTGGCTGCGCTTATAGTAAAACATGGCGAACAAAAAGAAGCATTTCTGAAGGCCTGCACTCTCGCTCGCCGAACAGCCGAAACATTCTTAAAGTACGCTGGACGTTCGGCTCAAGTTCACGGCCAGACTCCAGCCGCAAGCAAGGCTCATCATGAGCAGACACGAGCTATACTTGACACCTTATTGGCCCAAGAAACAAAGGTATTGGAACATTGGACGGTACGCAAGAAGCGTCTAGAACAATGTCAGCAGTTTGCTTTATTCGAGCGTTCGGCCCGCGCCGCAGTCGAGTGGATCCGTGAGACTCAGGAACGCTGCGGAGCTGCCGCGGCCGCTGCTGCTGCGGGCGTCGCTAGGGAGAGTCGCGAGCGAGTGCGTTTGCTCGCTCAGTTGGCAGACGGACTAGTCGAGAAGGGACACCCTCATGCCGTTCAGATTAAAGAATGGGTTGCTGCTGTTGACGCCAGATACGCGGAATTCAGTGGTTCTATGGAAGGTGGAGAGAGCGAACCAGAAAGTGACTCAGGCGTGGCTCCATCCTTAGCATCATCGCAATCTGCTGATAACGACAACCGTGCTGAACCACAGCCACCAACTATAGGCGATGATAAACGGCGTAGCGCCAGACGTAAAGAGTTTATAATGGCAGAGCTGCTACAAACTGAACGTGCCTACGTGAAAGATCTGGAGACCTGCATTACTTGTTATTTGCGTGAAATGCGTACCGATCCCGCCTCCGTACCGCTGCCACTTCAAGGAAAGGAGGACTTGATATTTGGTAACATAGAAGAGATTCATCGTTTCCACGAGCGCGTTTTCCTCAGAGAATTGGACAAATACGAGACGATGCCGGAAGACGTCGGCCACTGCTTCGTGACTTGGGCGCGCGAGTTCGATATGTACGTGTCGTACTGTCGCAACAAGCCAGACAGCAATGCGGCTGTCGTCCAACACGCCGGCGATTACTTTGAGAGGGTGCAACGAAAGAAAAAACTCGAGCATCCACTCGCCGCTTATTTGATCAAACCGGTTCAAAGGATTACCAAGTATCAACTGCTGCTGAAGGATCTTCAGGCATGCTGTGCTGAAGGACAAGGAGAGATAAAGGACGGCTTGGAAGTTATGCTCTCAGTGCCAAAGAAGGCCAATGATGCGATGCACCTCTCTCTGCTCGAGGGGTGTGACGTCCCGACTGACAGCCTCGGCGAGGTCGTCCTCCAGGACTCCTTCCACGTGTGGGACCTTCGTCAGATCATCAAAAAGGGACGCGAGAGGAGAGTCTTCCTTTTCGACTTGCATCTGCTTTTGGCGAAAGAAGTGAAGGATTCGCACGGAAAAGCCAAATACATTTACAAGACTAAATTTATGACATCAGAACTTGGGGTGACAGAACACATCGAAGGAGACGAATGCAAATTCTCGGTGTGGACTGGCCGCGAGCCAATGGCCAGCGACTGTCGCATCGTACTTAAGGCACCCTCGCTTGAAGTGAAGCAAACTTGGGTACGGAGTCTCAGGGAGGTTATACAGGAAACTTATTTCAGCTGTGCCCTGCCTCCCAGGAGCCCTGCACGGGTTCGCCCTACAAGCTCTCAACGATCTAGCAGGGATTTCGAAGACACCGTACTAGACGAAACGACGGAAAATCTCGACAGAAACTCGCTCGCGTCTTTCGGCAGTGGTAATACCACGGATTCCGATAAGGCCGGACCAGAAATGACATGGGTGATTTATGAGCACACCTCGGGTGGAACTGGTGAACTAACTGTGACAAAAGGCCAGCAAGTGGAGGTGGTAGAACCGTGGACCACTCGGCCTGACTGGTGGGTGGTCCGGGTTCCAGGGGAGCCACCACTAGAAGGTGCAGTACCAGCTCAGGTGCTCAAGCCGCAGCCACAACATCCGCCACATAAGACATCACCCTCCAGACGTCCTCTAAGCCAACCCTGTGAGGAAACGCTAGGGTTGTAA
Protein
MEAGARALDVLPLLQERVASLTGGRDRRGSPIIWFPANSRRDRVAPDDYRRLIHYLISIPGESVKSLGFTILIDMRGSAWAAIKPILKVLQEHFPSSVHQALVVKPDNFWQKQRTTIGAHKYKFETTMISLEALPKVIDSSQLTPDLEGTLQYDHPQWIDLRLALEELMWQAGELLDRLDDLQEDVARADFADDVTGARRAIDAHADISKRLAKVPVDELEAQGERVLQRLEVAEAACSEASGSGAEGSAFHCGSPGAVRAQLSAVRSAHAHAHKLWQHKKMQLDQCFQLRLFEQDCEKMLEWIVNHRTAFLATYVEIGRSCAAAKRLQEEHARFAAACTSGGRPSVARVTTAARRLADKRHYAESQISALAQRLERAYKQLAAGLEERSAVLSLSVVFHHKAEAYASAVGGWGAQCAVALQLATGQGGAPCNDPRALEQHAQRHRQLYEHMCQAYTEVHSTSKKLLYQLDHLVQVCHQTDPTDMQSEGTVSSAGSSGGDPAADYSSGANHVLAVIHQILGHHRALEARYHQARLKLHQRLALLLYKEDCRQVLDWLANHGEVFLQKNTGIGRNLHKARVYQKSHEHFENVAQNTYTNAEKLLGAAEELARSGECDPEEVLGVARELEAHVAAFAARVDRRRRRLDLAVLLYTHEKELLQWLDTLRSGGGVCATDDTPEAARRALEQCAQHRAASMDACAATIAQGEALMQDLRSSGDSDATSTAAVESTLERLRGTRSALEELWSDRERKLELTLQLRHFERDALEVSSRLELWGDELQRTEPPRDPQQAEQALAAHNESVARMQHATFQVVQQGQELAAAIADASDVMASSGSDGTETAPLDAQGRVQLLLEFLHDRQLDLEELAEERRARFEQCVQLGQFQKDAAQVVSWIRNGEAMLAASFSIPGTLSEAEQLKREHDQFQVAVEKTHASAVQVKYRADALRAANHYDPHTIREISEEVTERWQRLVTCAEERHKLVTASLNFYKTAEQVCSVLDSLEREYRRDEDWCGSAAAPSTDEPTLAQSIDKAAQVAALIVKHGEQKEAFLKACTLARRTAETFLKYAGRSAQVHGQTPAASKAHHEQTRAILDTLLAQETKVLEHWTVRKKRLEQCQQFALFERSARAAVEWIRETQERCGAAAAAAAAGVARESRERVRLLAQLADGLVEKGHPHAVQIKEWVAAVDARYAEFSGSMEGGESEPESDSGVAPSLASSQSADNDNRAEPQPPTIGDDKRRSARRKEFIMAELLQTERAYVKDLETCITCYLREMRTDPASVPLPLQGKEDLIFGNIEEIHRFHERVFLRELDKYETMPEDVGHCFVTWAREFDMYVSYCRNKPDSNAAVVQHAGDYFERVQRKKKLEHPLAAYLIKPVQRITKYQLLLKDLQACCAEGQGEIKDGLEVMLSVPKKANDAMHLSLLEGCDVPTDSLGEVVLQDSFHVWDLRQIIKKGRERRVFLFDLHLLLAKEVKDSHGKAKYIYKTKFMTSELGVTEHIEGDECKFSVWTGREPMASDCRIVLKAPSLEVKQTWVRSLREVIQETYFSCALPPRSPARVRPTSSQRSSRDFEDTVLDETTENLDRNSLASFGSGNTTDSDKAGPEMTWVIYEHTSGGTGELTVTKGQQVEVVEPWTTRPDWWVVRVPGEPPLEGAVPAQVLKPQPQHPPHKTSPSRRPLSQPCEETLGL
Summary
Uniprot
A0A139WLV9
A0A1Y1LVJ0
A0A1Y1LN33
A0A1Y1LT27
A0A1B6MEN6
A0A1B6KY89
+ More
A0A224XHF4 D6WFP9 A0A023F4M4 A0A1B6D8H9 A0A1B6C0A3 A0A139WM15 N6T7E4 U4TVJ0 A0A154NYC3 A0A0A1X1Q9 A0A0K8U2P5 A0A0L7QSK7 W8B4T1 A0A3L8DNR6 A0A2A3EIZ4 A0A034VK66 A0A1A9VYL1 A0A1A9Y9M6 A0A1A9Z3D8 A0A0L0CBX6 A0A1B0B9B4 B4KWS0 J9JLT4 A0A146KZ29 A0A158NJF7 A0A1S4EYM5 A0A151WI51 A0A151IDZ8 W8AIE7 E0VF59 A0A1A9X2W8 T1IXB4 A0A131XYD0 A0A1B0CFL6 A0A1B0CVW0 L7LZP7 A0A131YCL9 A0A131Z2U8 A0A131Z835 A0A2R5L7A1 L7MMH5 L7MKT8 A0A1E1X333 A0A1B6LW40 A0A1B6L258 A0A1S3J8Q8 A0A2T7PZK7 A0A1S3J9Q9 A0A1S3J882 A0A1S3J8G6 A0A1S3J887 A0A1S3J9Q4 A0A1S3J879 A0A1S3J8H2 A0A067RDN4 H3CV36 A0A3P8RS19 A0A3B4WWY5 A0A3B4ZDR0 A0A3B4TAE6 A0A3P8PXA1 A0A3P9CP01 A0A3B4FXU3 A0A3B3XN93 A0A2U9BLC7 A0A091J9E6 I3JLR7 A0A2D0PUW6 A0A093I4L9 A0A1A7ZL21 W5LXL5 H0Z1A3 A0A3Q0H079 A0A1D5P5L9 A0A1A8B1J2 A0A3Q0H081 A0A1W5ASA4 A0A3Q0H075 A0A3Q0H0Z8 A0A2I4BAC2 A0A3Q0H076 A0A146W7U3 A0A3Q0H4S7 A0A146W587 A0A091I4C1 A0A1A8BP83 A0A146NWQ9 A0A151M114 A0A1A8V7V5 A0A1V4JWH0
A0A224XHF4 D6WFP9 A0A023F4M4 A0A1B6D8H9 A0A1B6C0A3 A0A139WM15 N6T7E4 U4TVJ0 A0A154NYC3 A0A0A1X1Q9 A0A0K8U2P5 A0A0L7QSK7 W8B4T1 A0A3L8DNR6 A0A2A3EIZ4 A0A034VK66 A0A1A9VYL1 A0A1A9Y9M6 A0A1A9Z3D8 A0A0L0CBX6 A0A1B0B9B4 B4KWS0 J9JLT4 A0A146KZ29 A0A158NJF7 A0A1S4EYM5 A0A151WI51 A0A151IDZ8 W8AIE7 E0VF59 A0A1A9X2W8 T1IXB4 A0A131XYD0 A0A1B0CFL6 A0A1B0CVW0 L7LZP7 A0A131YCL9 A0A131Z2U8 A0A131Z835 A0A2R5L7A1 L7MMH5 L7MKT8 A0A1E1X333 A0A1B6LW40 A0A1B6L258 A0A1S3J8Q8 A0A2T7PZK7 A0A1S3J9Q9 A0A1S3J882 A0A1S3J8G6 A0A1S3J887 A0A1S3J9Q4 A0A1S3J879 A0A1S3J8H2 A0A067RDN4 H3CV36 A0A3P8RS19 A0A3B4WWY5 A0A3B4ZDR0 A0A3B4TAE6 A0A3P8PXA1 A0A3P9CP01 A0A3B4FXU3 A0A3B3XN93 A0A2U9BLC7 A0A091J9E6 I3JLR7 A0A2D0PUW6 A0A093I4L9 A0A1A7ZL21 W5LXL5 H0Z1A3 A0A3Q0H079 A0A1D5P5L9 A0A1A8B1J2 A0A3Q0H081 A0A1W5ASA4 A0A3Q0H075 A0A3Q0H0Z8 A0A2I4BAC2 A0A3Q0H076 A0A146W7U3 A0A3Q0H4S7 A0A146W587 A0A091I4C1 A0A1A8BP83 A0A146NWQ9 A0A151M114 A0A1A8V7V5 A0A1V4JWH0
Pubmed
EMBL
KQ971319
KYB28883.1
GEZM01051301
JAV75037.1
GEZM01051308
JAV75029.1
+ More
GEZM01051303 JAV75035.1 GEBQ01005618 JAT34359.1 GEBQ01023575 JAT16402.1 GFTR01008957 JAW07469.1 EFA00235.2 GBBI01002410 JAC16302.1 GEDC01015299 GEDC01006504 JAS21999.1 JAS30794.1 GEDC01030351 GEDC01012604 JAS06947.1 JAS24694.1 KYB28885.1 APGK01049651 APGK01049652 KB741156 ENN73578.1 KB631725 ERL85599.1 KQ434783 KZC04669.1 GBXI01009689 GBXI01007173 GBXI01002307 JAD04603.1 JAD07119.1 JAD11985.1 GDHF01031332 GDHF01022266 JAI20982.1 JAI30048.1 KQ414758 KOC61451.1 GAMC01018254 GAMC01018252 JAB88301.1 QOIP01000006 RLU21448.1 KZ288227 PBC31735.1 GAKP01016777 JAC42175.1 JRES01000611 KNC29898.1 JXJN01010338 JXJN01010339 JXJN01010340 CH933809 EDW17517.2 KRG05584.1 ABLF02035913 ABLF02035914 ABLF02035926 ABLF02035927 ABLF02035933 ABLF02035935 ABLF02035937 ABLF02035938 ABLF02035939 ABLF02060898 GDHC01017005 GDHC01007329 JAQ01624.1 JAQ11300.1 ADTU01017677 ADTU01017678 ADTU01017679 ADTU01017680 ADTU01017681 KQ983097 KYQ47508.1 KQ977901 KYM98901.1 GAMC01018253 GAMC01018250 JAB88305.1 DS235101 EEB12015.1 JH431646 GEFM01004538 JAP71258.1 AJWK01010177 AJWK01010178 AJWK01010179 AJWK01031337 GACK01008705 JAA56329.1 GEDV01011483 JAP77074.1 GEDV01003487 JAP85070.1 GEDV01001565 JAP86992.1 GGLE01001242 MBY05368.1 GACK01000475 JAA64559.1 GACK01000474 JAA64560.1 GFAC01005742 JAT93446.1 GEBQ01012080 JAT27897.1 GEBQ01022190 JAT17787.1 PZQS01000001 PVD38852.1 KK852768 KDR16954.1 CP026249 AWP04894.1 KK501769 KFP17644.1 AERX01010801 AERX01010802 KL206896 KFV86790.1 HADY01004663 SBP43148.1 AHAT01012339 ABQF01011024 ABQF01011025 ABQF01011026 ABQF01011027 AADN05000350 HADY01021860 SBP60345.1 GCES01061174 JAR25149.1 GCES01061175 JAR25148.1 KL218243 KFP03374.1 HADZ01004722 SBP68663.1 GCES01150143 JAQ36179.1 AKHW03006853 KYO18170.1 HAEJ01014851 SBS55308.1 LSYS01005643 OPJ76494.1
GEZM01051303 JAV75035.1 GEBQ01005618 JAT34359.1 GEBQ01023575 JAT16402.1 GFTR01008957 JAW07469.1 EFA00235.2 GBBI01002410 JAC16302.1 GEDC01015299 GEDC01006504 JAS21999.1 JAS30794.1 GEDC01030351 GEDC01012604 JAS06947.1 JAS24694.1 KYB28885.1 APGK01049651 APGK01049652 KB741156 ENN73578.1 KB631725 ERL85599.1 KQ434783 KZC04669.1 GBXI01009689 GBXI01007173 GBXI01002307 JAD04603.1 JAD07119.1 JAD11985.1 GDHF01031332 GDHF01022266 JAI20982.1 JAI30048.1 KQ414758 KOC61451.1 GAMC01018254 GAMC01018252 JAB88301.1 QOIP01000006 RLU21448.1 KZ288227 PBC31735.1 GAKP01016777 JAC42175.1 JRES01000611 KNC29898.1 JXJN01010338 JXJN01010339 JXJN01010340 CH933809 EDW17517.2 KRG05584.1 ABLF02035913 ABLF02035914 ABLF02035926 ABLF02035927 ABLF02035933 ABLF02035935 ABLF02035937 ABLF02035938 ABLF02035939 ABLF02060898 GDHC01017005 GDHC01007329 JAQ01624.1 JAQ11300.1 ADTU01017677 ADTU01017678 ADTU01017679 ADTU01017680 ADTU01017681 KQ983097 KYQ47508.1 KQ977901 KYM98901.1 GAMC01018253 GAMC01018250 JAB88305.1 DS235101 EEB12015.1 JH431646 GEFM01004538 JAP71258.1 AJWK01010177 AJWK01010178 AJWK01010179 AJWK01031337 GACK01008705 JAA56329.1 GEDV01011483 JAP77074.1 GEDV01003487 JAP85070.1 GEDV01001565 JAP86992.1 GGLE01001242 MBY05368.1 GACK01000475 JAA64559.1 GACK01000474 JAA64560.1 GFAC01005742 JAT93446.1 GEBQ01012080 JAT27897.1 GEBQ01022190 JAT17787.1 PZQS01000001 PVD38852.1 KK852768 KDR16954.1 CP026249 AWP04894.1 KK501769 KFP17644.1 AERX01010801 AERX01010802 KL206896 KFV86790.1 HADY01004663 SBP43148.1 AHAT01012339 ABQF01011024 ABQF01011025 ABQF01011026 ABQF01011027 AADN05000350 HADY01021860 SBP60345.1 GCES01061174 JAR25149.1 GCES01061175 JAR25148.1 KL218243 KFP03374.1 HADZ01004722 SBP68663.1 GCES01150143 JAQ36179.1 AKHW03006853 KYO18170.1 HAEJ01014851 SBS55308.1 LSYS01005643 OPJ76494.1
Proteomes
UP000007266
UP000019118
UP000030742
UP000076502
UP000053825
UP000279307
+ More
UP000242457 UP000078200 UP000092443 UP000092445 UP000037069 UP000092460 UP000009192 UP000007819 UP000005205 UP000075809 UP000078542 UP000009046 UP000091820 UP000092461 UP000085678 UP000245119 UP000027135 UP000007303 UP000265080 UP000261360 UP000261400 UP000261420 UP000265100 UP000265160 UP000261460 UP000261480 UP000246464 UP000053119 UP000005207 UP000221080 UP000053584 UP000018468 UP000007754 UP000189705 UP000000539 UP000192224 UP000192220 UP000054308 UP000050525 UP000190648
UP000242457 UP000078200 UP000092443 UP000092445 UP000037069 UP000092460 UP000009192 UP000007819 UP000005205 UP000075809 UP000078542 UP000009046 UP000091820 UP000092461 UP000085678 UP000245119 UP000027135 UP000007303 UP000265080 UP000261360 UP000261400 UP000261420 UP000265100 UP000265160 UP000261460 UP000261480 UP000246464 UP000053119 UP000005207 UP000221080 UP000053584 UP000018468 UP000007754 UP000189705 UP000000539 UP000192224 UP000192220 UP000054308 UP000050525 UP000190648
Pfam
Interpro
IPR001251
CRAL-TRIO_dom
+ More
IPR001331 GDS_CDC24_CS
IPR011993 PH-like_dom_sf
IPR002017 Spectrin_repeat
IPR036028 SH3-like_dom_sf
IPR001849 PH_domain
IPR000219 DH-domain
IPR018159 Spectrin/alpha-actinin
IPR001452 SH3_domain
IPR035899 DBL_dom_sf
IPR028570 TRIO
IPR036865 CRAL-TRIO_dom_sf
IPR028569 Kalirin
IPR013783 Ig-like_fold
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
IPR008271 Ser/Thr_kinase_AS
IPR036116 FN3_sf
IPR013098 Ig_I-set
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR036179 Ig-like_dom_sf
IPR003006 Ig/MHC_CS
IPR017441 Protein_kinase_ATP_BS
IPR003598 Ig_sub2
IPR001331 GDS_CDC24_CS
IPR011993 PH-like_dom_sf
IPR002017 Spectrin_repeat
IPR036028 SH3-like_dom_sf
IPR001849 PH_domain
IPR000219 DH-domain
IPR018159 Spectrin/alpha-actinin
IPR001452 SH3_domain
IPR035899 DBL_dom_sf
IPR028570 TRIO
IPR036865 CRAL-TRIO_dom_sf
IPR028569 Kalirin
IPR013783 Ig-like_fold
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
IPR008271 Ser/Thr_kinase_AS
IPR036116 FN3_sf
IPR013098 Ig_I-set
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR036179 Ig-like_dom_sf
IPR003006 Ig/MHC_CS
IPR017441 Protein_kinase_ATP_BS
IPR003598 Ig_sub2
SUPFAM
Gene 3D
ProteinModelPortal
A0A139WLV9
A0A1Y1LVJ0
A0A1Y1LN33
A0A1Y1LT27
A0A1B6MEN6
A0A1B6KY89
+ More
A0A224XHF4 D6WFP9 A0A023F4M4 A0A1B6D8H9 A0A1B6C0A3 A0A139WM15 N6T7E4 U4TVJ0 A0A154NYC3 A0A0A1X1Q9 A0A0K8U2P5 A0A0L7QSK7 W8B4T1 A0A3L8DNR6 A0A2A3EIZ4 A0A034VK66 A0A1A9VYL1 A0A1A9Y9M6 A0A1A9Z3D8 A0A0L0CBX6 A0A1B0B9B4 B4KWS0 J9JLT4 A0A146KZ29 A0A158NJF7 A0A1S4EYM5 A0A151WI51 A0A151IDZ8 W8AIE7 E0VF59 A0A1A9X2W8 T1IXB4 A0A131XYD0 A0A1B0CFL6 A0A1B0CVW0 L7LZP7 A0A131YCL9 A0A131Z2U8 A0A131Z835 A0A2R5L7A1 L7MMH5 L7MKT8 A0A1E1X333 A0A1B6LW40 A0A1B6L258 A0A1S3J8Q8 A0A2T7PZK7 A0A1S3J9Q9 A0A1S3J882 A0A1S3J8G6 A0A1S3J887 A0A1S3J9Q4 A0A1S3J879 A0A1S3J8H2 A0A067RDN4 H3CV36 A0A3P8RS19 A0A3B4WWY5 A0A3B4ZDR0 A0A3B4TAE6 A0A3P8PXA1 A0A3P9CP01 A0A3B4FXU3 A0A3B3XN93 A0A2U9BLC7 A0A091J9E6 I3JLR7 A0A2D0PUW6 A0A093I4L9 A0A1A7ZL21 W5LXL5 H0Z1A3 A0A3Q0H079 A0A1D5P5L9 A0A1A8B1J2 A0A3Q0H081 A0A1W5ASA4 A0A3Q0H075 A0A3Q0H0Z8 A0A2I4BAC2 A0A3Q0H076 A0A146W7U3 A0A3Q0H4S7 A0A146W587 A0A091I4C1 A0A1A8BP83 A0A146NWQ9 A0A151M114 A0A1A8V7V5 A0A1V4JWH0
A0A224XHF4 D6WFP9 A0A023F4M4 A0A1B6D8H9 A0A1B6C0A3 A0A139WM15 N6T7E4 U4TVJ0 A0A154NYC3 A0A0A1X1Q9 A0A0K8U2P5 A0A0L7QSK7 W8B4T1 A0A3L8DNR6 A0A2A3EIZ4 A0A034VK66 A0A1A9VYL1 A0A1A9Y9M6 A0A1A9Z3D8 A0A0L0CBX6 A0A1B0B9B4 B4KWS0 J9JLT4 A0A146KZ29 A0A158NJF7 A0A1S4EYM5 A0A151WI51 A0A151IDZ8 W8AIE7 E0VF59 A0A1A9X2W8 T1IXB4 A0A131XYD0 A0A1B0CFL6 A0A1B0CVW0 L7LZP7 A0A131YCL9 A0A131Z2U8 A0A131Z835 A0A2R5L7A1 L7MMH5 L7MKT8 A0A1E1X333 A0A1B6LW40 A0A1B6L258 A0A1S3J8Q8 A0A2T7PZK7 A0A1S3J9Q9 A0A1S3J882 A0A1S3J8G6 A0A1S3J887 A0A1S3J9Q4 A0A1S3J879 A0A1S3J8H2 A0A067RDN4 H3CV36 A0A3P8RS19 A0A3B4WWY5 A0A3B4ZDR0 A0A3B4TAE6 A0A3P8PXA1 A0A3P9CP01 A0A3B4FXU3 A0A3B3XN93 A0A2U9BLC7 A0A091J9E6 I3JLR7 A0A2D0PUW6 A0A093I4L9 A0A1A7ZL21 W5LXL5 H0Z1A3 A0A3Q0H079 A0A1D5P5L9 A0A1A8B1J2 A0A3Q0H081 A0A1W5ASA4 A0A3Q0H075 A0A3Q0H0Z8 A0A2I4BAC2 A0A3Q0H076 A0A146W7U3 A0A3Q0H4S7 A0A146W587 A0A091I4C1 A0A1A8BP83 A0A146NWQ9 A0A151M114 A0A1A8V7V5 A0A1V4JWH0
PDB
2NZ8
E-value=7.52321e-117,
Score=1082
Ontologies
GO
PANTHER
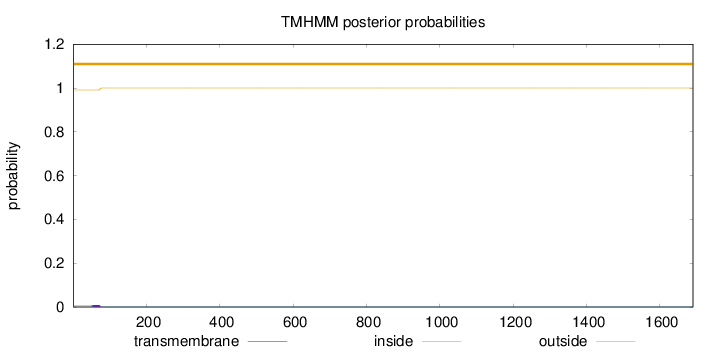
Topology
Length:
1690
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18764
Exp number, first 60 AAs:
0.07677
Total prob of N-in:
0.00840
outside
1 - 1690
Population Genetic Test Statistics
Pi
1.251147
Theta
15.178711
Tajima's D
-1.932636
CLR
0.141337
CSRT
0.016249187540623
Interpretation
Possibly Positive selection
