Gene
KWMTBOMO00545 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012336
Annotation
lactate_dehydrogenase_[Bombyx_mori]
Full name
L-lactate dehydrogenase
Location in the cell
Cytoplasmic Reliability : 1.359 Mitochondrial Reliability : 1.282
Sequence
CDS
ATGGCGACAAACAACACCAACGGGCATTGCGAGAACTACCACAAGATGGAGTCCCTGAAGAAGCTGTTCCAGCCCGTGCACGAGAAGGTGGACGAAACTTGGAGCAAGGTGACCATTGTCGGGGTCGGTCAGGTTGGGATGGCCGCAGCTTTCTCTATGCTGACGCAGAATGTTACGAATAACATCGCTCTAGTAGACATGATGGCTGACAAATTGAAAGGAGAGATGATGGACCTGCAGCACGGATCAGCATTCATGAGGAACGCCAAGATCCAATCTAGTACGGATTATTCTATAACAGCTGGCTCGAAGATCTGCGTGGTTACTGCTGGTGTTCGACAACGCGAAGGTGAATCTCGTCTCGATCTCGTGCAGAGAAACACCGATGTTCTTAAACAAATAATCCCGCAGCTGATAAAGTACAGTCCGGACACAATATTGGTGATCGCCAGTAACCCCGTGGATATTCTGACGTATGTTACGTGGAAGATTAGCGGGCTGCCCAAGCACCGCGTCATCGGGTCCGGCACTAACTTGGACTCGGCACGGTTCCGTTACCTGCTGTCGGACAGGCTCGGTATCGCTACCACGTCCTGCCACGGCTACATCATCGGCGAACACGGAGACAGCAGCGTTCCAGTATGGTCTGCAGTAAACATAGCGGGAGTACGCCTCAGTGATCTCAACAATCAGATCGGAACCGACGACGATCCTGAGAACTGGAAGGAGCTTCACGAGAATGTCGTGAAAAGTGCTTACGAAGTCATCAAACTGAAGGGGTACACTTCCTGGGCTATCGGACTGTCGCTGGCTCAGATCGTGAGGGCTATACTCACGAACGCTAACAGCGTTCACGCGGTCTCTACTTATCTCAAGGGCGAGCACGGCATCGAAGACGAAGTATTCTTGTCGCTGCCGTGCGTGTTGAGTCACTGTGGAGTCTCTGACGTCATCCGCCAGCCTCTAACCGAACTTGAAGTGGCACAGCTTCGGAAATCTGCTAAAGTCATGGCTAAAGTGCAGAACGACATTAAATTCTAA
Protein
MATNNTNGHCENYHKMESLKKLFQPVHEKVDETWSKVTIVGVGQVGMAAAFSMLTQNVTNNIALVDMMADKLKGEMMDLQHGSAFMRNAKIQSSTDYSITAGSKICVVTAGVRQREGESRLDLVQRNTDVLKQIIPQLIKYSPDTILVIASNPVDILTYVTWKISGLPKHRVIGSGTNLDSARFRYLLSDRLGIATTSCHGYIIGEHGDSSVPVWSAVNIAGVRLSDLNNQIGTDDDPENWKELHENVVKSAYEVIKLKGYTSWAIGLSLAQIVRAILTNANSVHAVSTYLKGEHGIEDEVFLSLPCVLSHCGVSDVIRQPLTELEVAQLRKSAKVMAKVQNDIKF
Summary
Catalytic Activity
(S)-lactate + NAD(+) = H(+) + NADH + pyruvate
Subunit
Homotetramer.
Similarity
Belongs to the LDH/MDH superfamily.
Belongs to the LDH/MDH superfamily. LDH family.
Belongs to the LDH/MDH superfamily. LDH family.
Keywords
Complete proteome
Cytoplasm
NAD
Oxidoreductase
Reference proteome
Feature
chain L-lactate dehydrogenase
Uniprot
A7LBQ0
A0A212EKH7
A0A194R3C8
A0A194PLZ3
A0A2Z5U750
A0A2A4K4S0
+ More
A0A1L8E0T1 A0A1B0GME5 A0A1W5YLL0 B0X5J3 A0A1Q3FSB9 A0A182XNL4 A0A1W5YLK3 A0A0M4F297 B0XBA2 W8C3Q7 A0A182PSY9 A0A0A1X778 A0A182NNJ5 A0A182UW01 A0A182LHZ5 Q7Q981 A0A182HXQ7 Q16ND1 A0A182Q983 A0A023ENR9 A0A0A1X8S0 E2AWN0 A0A1W5YLK5 B4KVP9 A0A182UF79 A0A182YBS7 A0A182RKJ0 A0A0N0BIH8 E2C744 A0A182WCU6 A0A182MMA4 A0A1Y1LEN4 A0A034WG20 A0A2M4AXA4 A0A034WAY2 A0A1A9WG64 T1DN51 A0A336MNZ3 B4PJN3 B4QJD3 A0A195BJ78 A0A0K8WAP3 B5RJQ0 Q95028 A0A0K8W7A9 B3NGH0 F4X535 B3M9Z0 A0A182G976 A0A1W4V9Z3 Q32KE6 B4MM53 A0A3B0KJ77 T1P9J0 Q29FH9 B4LI89 A0A0R3P2E0 B4HUT4 A0A067QM11 A0A2M4BUQ1 A0A0Q9WJ03 A0A0K8TLG7 A0A1I8PL33 A0A182JRF2 A0A182J9A7 A0A3L8E292 A0A0L0CPC7 A0A0C9RB25 A0A1J1IQY3 A0A1A9Z2V0 D3TMP6 A0A1A9UJ90 B4IXY2 A0A310SE24 A0A1B6GJH6 A0A1B0G927 A0A1A9XJJ1 A0A0K8TMX1 K7J616 A0A182GI23 A0A1B6EQX6 A0A1B6F8P4 A0A026W644 A0A0P5BJL1 A0A0P5PUP7 A0A0P5GHI2 A0A164LZ80 A0A0N8CUU8 A0A0P5K1S2 A0A0J7LAF5 A0A0P5M5W4 E0W1P5 B7XH74 C6L1J5
A0A1L8E0T1 A0A1B0GME5 A0A1W5YLL0 B0X5J3 A0A1Q3FSB9 A0A182XNL4 A0A1W5YLK3 A0A0M4F297 B0XBA2 W8C3Q7 A0A182PSY9 A0A0A1X778 A0A182NNJ5 A0A182UW01 A0A182LHZ5 Q7Q981 A0A182HXQ7 Q16ND1 A0A182Q983 A0A023ENR9 A0A0A1X8S0 E2AWN0 A0A1W5YLK5 B4KVP9 A0A182UF79 A0A182YBS7 A0A182RKJ0 A0A0N0BIH8 E2C744 A0A182WCU6 A0A182MMA4 A0A1Y1LEN4 A0A034WG20 A0A2M4AXA4 A0A034WAY2 A0A1A9WG64 T1DN51 A0A336MNZ3 B4PJN3 B4QJD3 A0A195BJ78 A0A0K8WAP3 B5RJQ0 Q95028 A0A0K8W7A9 B3NGH0 F4X535 B3M9Z0 A0A182G976 A0A1W4V9Z3 Q32KE6 B4MM53 A0A3B0KJ77 T1P9J0 Q29FH9 B4LI89 A0A0R3P2E0 B4HUT4 A0A067QM11 A0A2M4BUQ1 A0A0Q9WJ03 A0A0K8TLG7 A0A1I8PL33 A0A182JRF2 A0A182J9A7 A0A3L8E292 A0A0L0CPC7 A0A0C9RB25 A0A1J1IQY3 A0A1A9Z2V0 D3TMP6 A0A1A9UJ90 B4IXY2 A0A310SE24 A0A1B6GJH6 A0A1B0G927 A0A1A9XJJ1 A0A0K8TMX1 K7J616 A0A182GI23 A0A1B6EQX6 A0A1B6F8P4 A0A026W644 A0A0P5BJL1 A0A0P5PUP7 A0A0P5GHI2 A0A164LZ80 A0A0N8CUU8 A0A0P5K1S2 A0A0J7LAF5 A0A0P5M5W4 E0W1P5 B7XH74 C6L1J5
EC Number
1.1.1.27
Pubmed
19121390
20852941
22118469
26354079
24495485
25830018
+ More
20966253 12364791 14747013 17210077 17510324 24945155 20798317 17994087 18057021 25244985 28004739 25348373 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9094207 21719571 26483478 25315136 15632085 24845553 26369729 30249741 26108605 20353571 20075255 24508170 20566863 19647052 19892987
20966253 12364791 14747013 17210077 17510324 24945155 20798317 17994087 18057021 25244985 28004739 25348373 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9094207 21719571 26483478 25315136 15632085 24845553 26369729 30249741 26108605 20353571 20075255 24508170 20566863 19647052 19892987
EMBL
BABH01017107
EU000385
EU334850
ABS18410.1
ABY60854.1
AGBW02014274
+ More
OWR41965.1 KQ460953 KPJ10346.1 KQ459599 KPI94347.1 AP017491 BBB06759.1 NWSH01000183 PCG78662.1 GFDF01001777 JAV12307.1 AJVK01011430 AJVK01011431 KX147667 ARI45071.1 DS232382 EDS40930.1 GFDL01004703 JAV30342.1 KX147665 ARI45069.1 CP012525 ALC45207.1 DS232622 EDS44193.1 GAMC01005964 JAC00592.1 GBXI01007133 JAD07159.1 AAAB01008905 EAA09690.2 APCN01005337 CH477829 EAT35843.1 AXCN02001915 GAPW01002521 JAC11077.1 GBXI01007157 JAD07135.1 GL443346 EFN62194.1 KX147666 ARI45070.1 CH933809 EDW18423.1 KQ435729 KOX77688.1 GL453303 EFN76243.1 AXCM01011455 GEZM01057941 JAV72083.1 GAKP01006264 GAKP01006263 JAC52688.1 GGFK01011907 MBW45228.1 GAKP01006261 JAC52691.1 GAMD01003180 JAA98410.1 UFQT01001695 SSX31455.1 CM000159 EDW93632.1 CM000363 CM002912 EDX09442.1 KMY97913.1 KQ976464 KYM84496.1 GDHF01004394 JAI47920.1 BT044524 AE014296 ACH95298.1 AGB94169.1 U68038 GDHF01005589 JAI46725.1 CH954178 EDV51136.1 GL888687 EGI58429.1 CH902618 EDV41210.1 KPU79002.1 JXUM01048950 JXUM01048951 JXUM01048952 JXUM01048953 JXUM01048954 KQ561584 KXJ78096.1 BT023933 ABB36437.1 CH963847 EDW73062.1 OUUW01000009 SPP85151.1 KA644830 AFP59459.1 CH379067 EAL31276.1 CH940647 EDW69656.1 KRT07361.1 CH480817 EDW50705.1 KK853182 KDR10168.1 GGFJ01007530 MBW56671.1 KRF84499.1 GDAI01002602 JAI15001.1 QOIP01000001 RLU26756.1 JRES01000089 KNC34223.1 GBYB01005520 JAG75287.1 CVRI01000058 CRL02639.1 EZ422698 ADD18974.1 CH916366 EDV96503.1 KQ762833 OAD55417.1 GECZ01009267 GECZ01007192 JAS60502.1 JAS62577.1 CCAG010002964 GDAI01001909 JAI15694.1 JXUM01065106 JXUM01065107 KQ562330 KXJ76135.1 GECZ01029636 GECZ01012506 GECZ01012328 JAS40133.1 JAS57263.1 JAS57441.1 GECZ01023184 GECZ01022006 GECZ01003330 GECZ01002734 JAS46585.1 JAS47763.1 JAS66439.1 JAS67035.1 KK107388 EZA51478.1 GDIP01184048 JAJ39354.1 GDIQ01144089 JAL07637.1 GDIQ01240964 JAK10761.1 LRGB01003123 KZS04571.1 GDIP01096663 JAM07052.1 GDIQ01196068 JAK55657.1 LBMM01000085 KMR05025.1 GDIQ01184318 JAK67407.1 DS235873 EEB19627.1 AB477534 BAH11456.1 JL616405 AB472336 AEH58612.1 BAH86734.1
OWR41965.1 KQ460953 KPJ10346.1 KQ459599 KPI94347.1 AP017491 BBB06759.1 NWSH01000183 PCG78662.1 GFDF01001777 JAV12307.1 AJVK01011430 AJVK01011431 KX147667 ARI45071.1 DS232382 EDS40930.1 GFDL01004703 JAV30342.1 KX147665 ARI45069.1 CP012525 ALC45207.1 DS232622 EDS44193.1 GAMC01005964 JAC00592.1 GBXI01007133 JAD07159.1 AAAB01008905 EAA09690.2 APCN01005337 CH477829 EAT35843.1 AXCN02001915 GAPW01002521 JAC11077.1 GBXI01007157 JAD07135.1 GL443346 EFN62194.1 KX147666 ARI45070.1 CH933809 EDW18423.1 KQ435729 KOX77688.1 GL453303 EFN76243.1 AXCM01011455 GEZM01057941 JAV72083.1 GAKP01006264 GAKP01006263 JAC52688.1 GGFK01011907 MBW45228.1 GAKP01006261 JAC52691.1 GAMD01003180 JAA98410.1 UFQT01001695 SSX31455.1 CM000159 EDW93632.1 CM000363 CM002912 EDX09442.1 KMY97913.1 KQ976464 KYM84496.1 GDHF01004394 JAI47920.1 BT044524 AE014296 ACH95298.1 AGB94169.1 U68038 GDHF01005589 JAI46725.1 CH954178 EDV51136.1 GL888687 EGI58429.1 CH902618 EDV41210.1 KPU79002.1 JXUM01048950 JXUM01048951 JXUM01048952 JXUM01048953 JXUM01048954 KQ561584 KXJ78096.1 BT023933 ABB36437.1 CH963847 EDW73062.1 OUUW01000009 SPP85151.1 KA644830 AFP59459.1 CH379067 EAL31276.1 CH940647 EDW69656.1 KRT07361.1 CH480817 EDW50705.1 KK853182 KDR10168.1 GGFJ01007530 MBW56671.1 KRF84499.1 GDAI01002602 JAI15001.1 QOIP01000001 RLU26756.1 JRES01000089 KNC34223.1 GBYB01005520 JAG75287.1 CVRI01000058 CRL02639.1 EZ422698 ADD18974.1 CH916366 EDV96503.1 KQ762833 OAD55417.1 GECZ01009267 GECZ01007192 JAS60502.1 JAS62577.1 CCAG010002964 GDAI01001909 JAI15694.1 JXUM01065106 JXUM01065107 KQ562330 KXJ76135.1 GECZ01029636 GECZ01012506 GECZ01012328 JAS40133.1 JAS57263.1 JAS57441.1 GECZ01023184 GECZ01022006 GECZ01003330 GECZ01002734 JAS46585.1 JAS47763.1 JAS66439.1 JAS67035.1 KK107388 EZA51478.1 GDIP01184048 JAJ39354.1 GDIQ01144089 JAL07637.1 GDIQ01240964 JAK10761.1 LRGB01003123 KZS04571.1 GDIP01096663 JAM07052.1 GDIQ01196068 JAK55657.1 LBMM01000085 KMR05025.1 GDIQ01184318 JAK67407.1 DS235873 EEB19627.1 AB477534 BAH11456.1 JL616405 AB472336 AEH58612.1 BAH86734.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000092462
+ More
UP000002320 UP000076407 UP000092553 UP000075885 UP000075884 UP000075903 UP000075882 UP000007062 UP000075840 UP000008820 UP000075886 UP000000311 UP000009192 UP000075902 UP000076408 UP000075900 UP000053105 UP000008237 UP000075920 UP000075883 UP000091820 UP000002282 UP000000304 UP000078540 UP000000803 UP000008711 UP000007755 UP000007801 UP000069940 UP000249989 UP000192221 UP000007798 UP000268350 UP000095301 UP000001819 UP000008792 UP000001292 UP000027135 UP000095300 UP000075881 UP000075880 UP000279307 UP000037069 UP000183832 UP000092445 UP000078200 UP000001070 UP000092444 UP000092443 UP000002358 UP000053097 UP000076858 UP000036403 UP000009046 UP000002281
UP000002320 UP000076407 UP000092553 UP000075885 UP000075884 UP000075903 UP000075882 UP000007062 UP000075840 UP000008820 UP000075886 UP000000311 UP000009192 UP000075902 UP000076408 UP000075900 UP000053105 UP000008237 UP000075920 UP000075883 UP000091820 UP000002282 UP000000304 UP000078540 UP000000803 UP000008711 UP000007755 UP000007801 UP000069940 UP000249989 UP000192221 UP000007798 UP000268350 UP000095301 UP000001819 UP000008792 UP000001292 UP000027135 UP000095300 UP000075881 UP000075880 UP000279307 UP000037069 UP000183832 UP000092445 UP000078200 UP000001070 UP000092444 UP000092443 UP000002358 UP000053097 UP000076858 UP000036403 UP000009046 UP000002281
Interpro
Gene 3D
ProteinModelPortal
A7LBQ0
A0A212EKH7
A0A194R3C8
A0A194PLZ3
A0A2Z5U750
A0A2A4K4S0
+ More
A0A1L8E0T1 A0A1B0GME5 A0A1W5YLL0 B0X5J3 A0A1Q3FSB9 A0A182XNL4 A0A1W5YLK3 A0A0M4F297 B0XBA2 W8C3Q7 A0A182PSY9 A0A0A1X778 A0A182NNJ5 A0A182UW01 A0A182LHZ5 Q7Q981 A0A182HXQ7 Q16ND1 A0A182Q983 A0A023ENR9 A0A0A1X8S0 E2AWN0 A0A1W5YLK5 B4KVP9 A0A182UF79 A0A182YBS7 A0A182RKJ0 A0A0N0BIH8 E2C744 A0A182WCU6 A0A182MMA4 A0A1Y1LEN4 A0A034WG20 A0A2M4AXA4 A0A034WAY2 A0A1A9WG64 T1DN51 A0A336MNZ3 B4PJN3 B4QJD3 A0A195BJ78 A0A0K8WAP3 B5RJQ0 Q95028 A0A0K8W7A9 B3NGH0 F4X535 B3M9Z0 A0A182G976 A0A1W4V9Z3 Q32KE6 B4MM53 A0A3B0KJ77 T1P9J0 Q29FH9 B4LI89 A0A0R3P2E0 B4HUT4 A0A067QM11 A0A2M4BUQ1 A0A0Q9WJ03 A0A0K8TLG7 A0A1I8PL33 A0A182JRF2 A0A182J9A7 A0A3L8E292 A0A0L0CPC7 A0A0C9RB25 A0A1J1IQY3 A0A1A9Z2V0 D3TMP6 A0A1A9UJ90 B4IXY2 A0A310SE24 A0A1B6GJH6 A0A1B0G927 A0A1A9XJJ1 A0A0K8TMX1 K7J616 A0A182GI23 A0A1B6EQX6 A0A1B6F8P4 A0A026W644 A0A0P5BJL1 A0A0P5PUP7 A0A0P5GHI2 A0A164LZ80 A0A0N8CUU8 A0A0P5K1S2 A0A0J7LAF5 A0A0P5M5W4 E0W1P5 B7XH74 C6L1J5
A0A1L8E0T1 A0A1B0GME5 A0A1W5YLL0 B0X5J3 A0A1Q3FSB9 A0A182XNL4 A0A1W5YLK3 A0A0M4F297 B0XBA2 W8C3Q7 A0A182PSY9 A0A0A1X778 A0A182NNJ5 A0A182UW01 A0A182LHZ5 Q7Q981 A0A182HXQ7 Q16ND1 A0A182Q983 A0A023ENR9 A0A0A1X8S0 E2AWN0 A0A1W5YLK5 B4KVP9 A0A182UF79 A0A182YBS7 A0A182RKJ0 A0A0N0BIH8 E2C744 A0A182WCU6 A0A182MMA4 A0A1Y1LEN4 A0A034WG20 A0A2M4AXA4 A0A034WAY2 A0A1A9WG64 T1DN51 A0A336MNZ3 B4PJN3 B4QJD3 A0A195BJ78 A0A0K8WAP3 B5RJQ0 Q95028 A0A0K8W7A9 B3NGH0 F4X535 B3M9Z0 A0A182G976 A0A1W4V9Z3 Q32KE6 B4MM53 A0A3B0KJ77 T1P9J0 Q29FH9 B4LI89 A0A0R3P2E0 B4HUT4 A0A067QM11 A0A2M4BUQ1 A0A0Q9WJ03 A0A0K8TLG7 A0A1I8PL33 A0A182JRF2 A0A182J9A7 A0A3L8E292 A0A0L0CPC7 A0A0C9RB25 A0A1J1IQY3 A0A1A9Z2V0 D3TMP6 A0A1A9UJ90 B4IXY2 A0A310SE24 A0A1B6GJH6 A0A1B0G927 A0A1A9XJJ1 A0A0K8TMX1 K7J616 A0A182GI23 A0A1B6EQX6 A0A1B6F8P4 A0A026W644 A0A0P5BJL1 A0A0P5PUP7 A0A0P5GHI2 A0A164LZ80 A0A0N8CUU8 A0A0P5K1S2 A0A0J7LAF5 A0A0P5M5W4 E0W1P5 B7XH74 C6L1J5
PDB
1T2F
E-value=1.1313e-120,
Score=1108
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
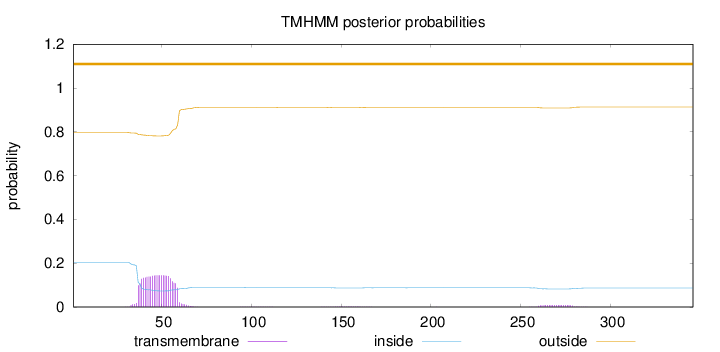
Topology
Subcellular location
Cytoplasm
Length:
346
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.36660000000001
Exp number, first 60 AAs:
3.08462
Total prob of N-in:
0.20349
outside
1 - 346
Population Genetic Test Statistics
Pi
23.826275
Theta
24.807229
Tajima's D
0.006771
CLR
0.023639
CSRT
0.375431228438578
Interpretation
Uncertain
